Prognostic value of circadian rhythm-associated genes in breast cancer
- PMID: 40369575
- PMCID: PMC12077051
- DOI: 10.1186/s12957-025-03829-8
Prognostic value of circadian rhythm-associated genes in breast cancer
Abstract
Objective: Breast cancer (BC) remains the most prevalent malignancy among women. Clinical evidence indicates that genetic variations related to circadian rhythms, as well as the timing of therapeutic interventions, influence the response to radiation therapy and the toxicity of pharmacological treatments in women with BC. This study aimed to identify key circadian rhythm-related genes (CRGs) using bioinformatics and machine learning, and construct a prognostic model to predict clinical outcomes.
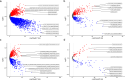
Methods: Transcriptome data for BC were retrieved from The Cancer Genome Atlas database. Univariate Cox regression and least absolute shrinkage and selection operator regression analyses were used to develop a prognostic model based on CRGs. The predictive performance of the risk score model was evaluated. Univariate and multivariate Cox regression analyses were applied to construct the prognostic model and stratify patients into high-risk and low-risk groups. Additionally, differences in immune microenvironment, immunotherapy efficacy, and tumor mutation burden were assessed between risk groups.
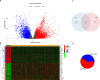
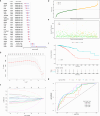
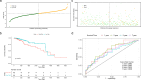
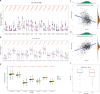
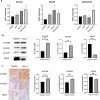
Results: A prognostic risk score model comprising 17 CRGs was developed. The areas under the receiver operating characteristic curve for overall survival at 1, 3, 5, and 7 years exceeded 0.6, indicating acceptable predictive performance. Calibration plots and decision curve analyses demonstrated the use of the model in prognostic prediction. Significant differences in immune microenvironment, immunotherapy efficacy, and tumor mutation burden were identified between the low-risk and high-risk groups.
Conclusion: The circadian rhythm-based gene model, effectively predicted the prognosis of individuals with BC, highlighting its potential to inform personalized therapeutic strategies and improve patient outcomes.
Keywords: Bioinformatic analysis; Breast cancer; Immune microenvironment; Prognosis; TMB.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study was conducted with approval from the Ethics Committee of Dongzhimen Hospital, Beijing University of Chinese Medicine. This study was conducted in accordance with the declaration of Helsinki. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures





Similar articles
-
Identification of cuproptosis-related subtypes, construction of a prognosis model, and tumor microenvironment landscape in gastric cancer.Front Immunol. 2022 Nov 21;13:1056932. doi: 10.3389/fimmu.2022.1056932. eCollection 2022. Front Immunol. 2022. PMID: 36479114 Free PMC article.
-
Machine learning-based diagnostic and prognostic models for breast cancer: a new frontier on the clinical application of natural killer cell-related gene signatures in precision medicine.Front Immunol. 2025 May 27;16:1581982. doi: 10.3389/fimmu.2025.1581982. eCollection 2025. Front Immunol. 2025. PMID: 40496857 Free PMC article.
-
A Circadian Rhythm-related Signature to Predict Prognosis, Immune Infiltration, and Drug Response in Breast Cancer.Curr Med Chem. 2025;32(3):608-626. doi: 10.2174/0109298673320179240803071001. Curr Med Chem. 2025. PMID: 39279697
-
Identifying novel circadian rhythm biomarkers for diagnosis and prognosis of melanoma by an integrated bioinformatics and machine learning approach.Aging (Albany NY). 2024 Jun 20;16(16):11824-11842. doi: 10.18632/aging.205961. Epub 2024 Jun 20. Aging (Albany NY). 2024. PMID: 39213172 Free PMC article.
-
Immunometabolism characteristics and a potential prognostic risk model associated with TP53 mutations in breast cancer.Front Immunol. 2022 Jul 22;13:946468. doi: 10.3389/fimmu.2022.946468. eCollection 2022. Front Immunol. 2022. PMID: 35935965 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

