Characterization of the transcriptional cellular response in midgut tissue of temephos-resistant Aedes aegypti larvae
- PMID: 40369600
- PMCID: PMC12076995
- DOI: 10.1186/s13071-025-06675-5
Characterization of the transcriptional cellular response in midgut tissue of temephos-resistant Aedes aegypti larvae
Abstract
Background: Resistance to organophosphate compounds is a serious concern in dealing with the control of mosquito vectors. Understanding the genetic and molecular basis of resistance is important not only to create strategies aimed at detecting and monitoring resistance in the field but also to implement efficient control measures and support the development of new insecticides. Despite the extensive literature on insecticide resistance, the molecular basis of metabolic resistance is still poorly understood.
Methods: To better understand the mechanisms of Aedes aegypti resistance to temephos, we performed high-throughput sequencing of RNA from the midgut tissue of Aedes aegypti larvae from a temephos-resistant laboratory colony, with long-term and continuous exposure to this insecticide (RecR), as well as from a reference, temephos-susceptible, colony (RecL). Bioinformatic analyses were then performed to assess the biological functions of differentially expressed genes, and the sequencing data were validated by quantitative reverse transcription-polymerase chain reaction (RT-qPCR).
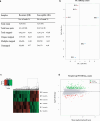
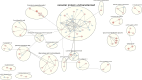
Results: The transcriptome analysis mapped 6.084 genes, of which 202 were considered upregulated in RecR, including known and new genes representing many detoxification enzyme families, such as cytochrome-P450 oxidative enzymes, glutathione-S-transferases and glucosyl transferases. Other upregulated genes were mainly involved in the cuticle, carbohydrates and lipid biosynthesis. For the downregulated profiles, we found 106 downregulated genes in the RecR colony, with molecules involved in protein synthesis, immunity and apoptosis process. Furthermore, we observed an enrichment of KEGG metabolic pathways related to resistance mechanisms. The results found in RT-qPCR confirm the findings of the transcriptome data.
Conclusions: In this study, we investigated transcriptome-level changes maintained in a temephos-resistant Ae. aegypti colony under continuous and prolonged selection pressure. Our results indicate that metabolic resistance might involve a larger and more significant number of detoxification enzymes, with different functional roles, than previously shown with other mechanisms, also contributing to the resistance phenotype in the Ae. aegypti RecR colony.
Keywords: Aedes aegypti; Metabolic resistance; Temephos; Transcriptome.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

