Intestinal microbiota profiles of captive-bred cynomolgus macaques reveal influence of biogeography and age
- PMID: 40369669
- PMCID: PMC12080069
- DOI: 10.1186/s42523-025-00409-9
Intestinal microbiota profiles of captive-bred cynomolgus macaques reveal influence of biogeography and age
Abstract
Background: Age-associated changes to the intestinal microbiome may be linked to inflammageing and the development of age-related chronic diseases. Cynomolgus macaques, a common animal model in biomedical research, have strong genetic physiological similarities to humans and may serve as beneficial models for the effect of age on the human microbiome. However, age-associated changes to their intestinal microbiome have previously only been investigated in faecal samples. Here, we have characterised and investigated the effects of age in the cynomolgus macaque intestinal tract in luminal samples from both the small and large intestine.
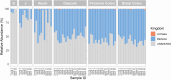
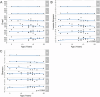
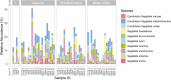
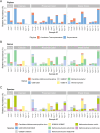
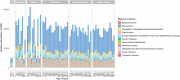
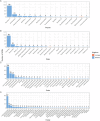
Results: Whole metagenomic shotgun sequencing was used to analyse the microbial communities in intestinal content obtained from six different intestinal regions, covering the duodenum to distal colon, of 24 healthy, captive-bred cynomolgus macaques, ranging in age from 4 to 20 years. Both reference-based and assembly-based computational profiling approaches were used to analyse changes to intestinal microbiota composition and metabolic potential associated with intestinal biogeography and age. Reference-based computational profiling revealed a significant and progressive increase in both species richness and evenness along the intestinal tract. The microbial community composition also significantly differed between the small intestine, caecum, and colon. Notably, no significant changes in the taxonomic abundance of individual taxa with age were found except when sex was included as a covariate. Additionally, using an assembly-based computational profiling approach, 156 putative novel bacterial and archaeal species were identified.
Conclusions: We observed limited effects of age on the composition of the luminal microbiota in the profiled regions of the intestinal tract except when sex was included as a covariate. The enteric microbial communities of the small and the large intestine were, however, distinct, highlighting the limitations of frequently used faecal microbial profiling as a proxy for the intestinal microbiota. The identification of a number of putative novel microbial taxa contributes to knowledge of the full diversity of the cynomolgus macaque intestinal microbiome.
Keywords: Ageing; Assembly-based computational profiling.; Cynomolgus macaque; Intestinal biogeography; Metagenomics; Microbiome; Non-human primate; Reference-based computational profiling.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures



Similar articles
-
Genetic hypogonadal mouse model reveals niche-specific influence of reproductive axis and sex on intestinal microbial communities.Biol Sex Differ. 2023 Nov 6;14(1):79. doi: 10.1186/s13293-023-00564-1. Biol Sex Differ. 2023. PMID: 37932822 Free PMC article.
-
The Cynomolgus Macaque Intestinal Mycobiome Is Dominated by the Kazachstania Genus and K. pintolopesii Species.J Fungi (Basel). 2022 Oct 8;8(10):1054. doi: 10.3390/jof8101054. J Fungi (Basel). 2022. PMID: 36294619 Free PMC article.
-
Gastrointestinal Biogeography of Luminal Microbiota and Short-Chain Fatty Acids in Sika Deer (Cervus nippon).Appl Environ Microbiol. 2022 Sep 13;88(17):e0049922. doi: 10.1128/aem.00499-22. Epub 2022 Aug 11. Appl Environ Microbiol. 2022. PMID: 35950850 Free PMC article.
-
Biogeography of intestinal mucus-associated microbiome: Depletion of genus Pseudomonas is associated with depressive-like behaviors in female cynomolgus macaques.J Adv Res. 2025 Apr;70:393-404. doi: 10.1016/j.jare.2024.05.013. Epub 2024 May 11. J Adv Res. 2025. PMID: 38735389 Free PMC article.
-
Metagenomic and network analysis revealed wide distribution of antibiotic resistance genes in monkey gut microbiota.Microbiol Res. 2022 Jan;254:126895. doi: 10.1016/j.micres.2021.126895. Epub 2021 Oct 23. Microbiol Res. 2022. PMID: 34742104 Review.
References
-
- Sender R, Fuchs S, Milo R. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell. 2016;164(3):337–40. 10.1016/j.cell.2016.01.013. - PubMed
-
- Martin-Gallausiaux C, Marinelli L, Blottiere HM, Larraufie P, Lapaque N. SCFA: mechanisms and functional importance in the gut. Proc Nutr Soc. 2021;80(1):37–49. 10.1017/S0029665120006916. - PubMed
-
- Venkatesh M, Mukherjee S, Wang H, Li H, Sun K, Benechet AP, Qiu Z, Maher L, Redinbo MR, Phillips RS, Fleet JC, Kortagere S, Mukherjee P, Fasano A, Le Ven J, Nicholson JK, Dumas ME, Khanna KM, Mani S. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity. 2014;41(2):296–310. 10.1016/j.immuni.2014.06.014. - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
