Parkin inhibits iron overload-induced cardiomyocyte ferroptosis by ubiquitinating ACSL4 and modulating PUFA-phospholipids metabolism
- PMID: 40370554
- PMCID: PMC12069115
- DOI: 10.1016/j.apsb.2024.12.027
Parkin inhibits iron overload-induced cardiomyocyte ferroptosis by ubiquitinating ACSL4 and modulating PUFA-phospholipids metabolism
Abstract
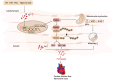
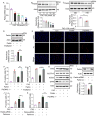
Iron overload is strongly associated with heart disease. Ferroptosis is a new form of regulated cell death indicated in cardiac ischemia-reperfusion (I/R) injury. However, the specific molecular mechanism of myocardial injury caused by iron overload in the heart is still unclear, and the involvement of ferroptosis in iron overload-induced myocardial injury is not fully understood. In this study, we observed that ferroptosis participated in developing of iron overload and I/R-induced cardiomyopathy. Mechanistically, we discovered that Parkin inhibited iron overload-induced ferroptosis in cardiomyocytes by promoting the ubiquitination of long-chain acyl-CoA synthetase 4 (ACSL4), a crucial protein involved in ferroptosis-related lipid metabolism pathways. Additionally, we identified p53 as a transcription factor that transcriptionally suppressed Parkin expression in iron-overloaded cardiomyocytes, thereby regulating iron overload-induced ferroptosis. In animal studies, cardiac-specific Parkin knockout mice (Myh6-CreER T2 /Parkin fl/fl ) fed a high-iron diet presented more severe myocardial damage, and the high iron levels exacerbated myocardial I/R injury. However, the ferroptosis inhibitor Fer-1 significantly suppressed iron overload-induced ferroptosis and myocardial I/R injury. Moreover, Parkin effectively protected against impaired mitochondrial function and prevented iron overload-induced mitochondrial lipid peroxidation. These findings unveil a novel regulatory pathway involving p53-Parkin-ACSL4 in heart disease by inhibiting of ferroptosis.
Keywords: ACSL4; Ferroptosis; Heart diseases; I/R; Iron overload; Mitochondrial; Parkin; p53.
© 2025 The Authors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
-
- Fernández Real J.M., López Bermejo A., Ricart W. Cross-talk between iron metabolism and diabetes. Diabetes. 2002;51:2348–2354. - PubMed
-
- Zhang J., Song Y., Li Y., Lin H.B., Fang X. Iron homeostasis in the heart: molecular mechanisms and pharmacological implications. J Mol Cell Cardiol. 2023;174:15–24. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

