Expression and diagnostic values of ferroptosis-related genes in coronavirus-associated viral sepsis
- PMID: 40370730
- PMCID: PMC12074931
- DOI: 10.3389/fmed.2025.1496834
Expression and diagnostic values of ferroptosis-related genes in coronavirus-associated viral sepsis
Abstract
Aim: The aim of this study is to investigate the differential expression and diagnostic value of ferroptosis-related genes in coronavirus-associated viral sepsis.
Methods: This study was conducted in two sequential phases: (1) identification of differentially expressed genes through comprehensive analysis of the experimental dataset (GSE164805); and (2) clinical validation of the candidate molecular markers using both test set samples and clinical samples, followed by rigorous evaluation of their diagnostic performance. Firstly, the microchips associated with coronavirus-associated viral sepsis were retrieved from the GEO database, a public data platform of NCBI (National Center for Biotechnology Information), and differentially expressed genes (DEGs) were obtained through differential analysis. The identified DEGs were then intersected with the ferroptosis gene dataset to obtain ferroptosis-related DEGs. Subsequently, molecular signaling pathways of ferroptosis-related genes in coronavirus-associated viral sepsis were analyzed using gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis. CIBERSORT was employed to analyze immune cell infiltration in both the coronavirus-associated viral sepsis group and control group. Furthermore, a protein-protein interaction (PPI) network was constructed to identify hub genes involved in ferroptosis. Finally, the expression of ferroptosis hub genes in coronavirus-associated viral sepsis and its diagnostic value were analyzed in validation set GSE199816 and clinical case samples.
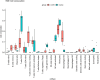
Results: In test set GSE164805, a total of 15,059 differentially expressed genes (DEGs) were identified, comprising 7,458 up-regulated and 7,601 down-regulated genes. Subsequently, an intersection analysis with the ferroptosis gene dataset yielded 189 DEGs associated with ferroptosis. Functional enrichment analyses using GO and KEGG revealed significant enrichment in signaling pathways related to ferroptosis, oxidative stress, and HIF-1. Additionally, CIBERSORT immune-infiltration analysis revealed enhanced infiltration of innate immune cells but reduced infiltration of CD8+ T cells and natural killer (NK) cells in the coronavirus-associated viral sepsis group compared with healthy controls. Furthermore, analysis identified that the distribution of these immune cells correlated with the expression levels of IL1-β and HMOX1, suggesting that viral infection in the septic pathological state disrupts the balance between immune activation and suppression. Notably, PPI network analysis also identified IL1-β and HMOX1 as hub genes involved in ferroptosis. Finally, the results were verified in the validation set and clinical case samples, and the results showed that the expressions of IL1-β and HMOX1 in the coronavirus-associated viral sepsis group were decreased compared with the case control group and the healthy control group. In clinical samples, the expression levels were as follows: IL1-β (0.390 ± 0.068 vs. 1.101 ± 0.107), HMOX1 (0.629 ± 0.117 vs. 1.101 ± 0.107), and the differences were statistically significant (all p < 0.05). Further diagnostic performance analysis demonstrated that IL1-β and HMOX1 exhibited AUROCs of 0.892 and 0.765, respectively, indicating their robust diagnostic potential for coronavirus-associated viral sepsis.
Conclusion: The present study has successfully identified two hub genes, IL1-β and HMOX1, associated with ferroptosis in coronavirus-associated viral sepsis, and their expression and diagnostic value for the disease. These findings provide effective diagnostic biomarkers and potential therapeutic targets for coronavirus-associated viral sepsis. Notably, this study specifically focused on coronavirus-induced viral sepsis, distinct from previously characterized bacterial sepsis and other viral etiologies, thus warranting future studies with expanded sample sizes for stratified analyses.
Keywords: HMOX1; IL1-β; coronavirus; ferroptosis; viral sepsis.
Copyright © 2025 Zhang, Wu and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures








References
-
- Fleischmann-Struzek C, Mellhammar L, Rose N, Cassini A, Rudd KE, Schlattmann P, et al. Incidence and mortality of hospital- and ICU-treated sepsis: results from an updated and expanded systematic review and meta-analysis. Intensive Care Med. (2020) 46:1552–62. doi: 10.1007/s00134-020-06151-x, PMID: - DOI - PMC - PubMed
-
- 徐逸天, 曹彬 . 病毒性感染中毒症——一个亟待重视的概念[J]. 中华结核和呼吸杂志. (2021) 44:674–9. doi: 10.3760/cma.j.cn112147-20201123-01111 - DOI
-
- van Houwelingen F, van Dellen E, Visser-Meily JMA, Valkenet K, Heijnen GH, Vernooij LM, et al. Mental, cognitive and physical outcomes after intensive care unit treatment during the COVID-19 pandemic: a comparison between COVID-19 and non-COVID-19 patients. Sci Rep. (2023) 13:14414. doi: 10.1038/s41598-023-41667-4, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

