A snapshot on molecular technologies for diagnosing FAdV infections
- PMID: 40370821
- PMCID: PMC12075531
- DOI: 10.3389/fvets.2025.1558257
A snapshot on molecular technologies for diagnosing FAdV infections
Abstract
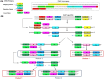
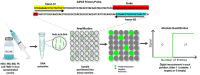
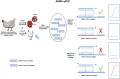
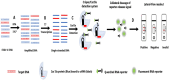
Fowl adenoviruses (FAdV) are prevalent in chickens worldwide, responsible for several poultry diseases, including inclusion body hepatitis (IBH), hepatitis-hydropericardium syndrome (HHS), and gizzard erosion (GE), which result in significant economic losses in the poultry industry. Consequently, detection and efficient identification of FAdV serotypes are becoming extremely urgent to monitor outbreaks and develop vaccination strategies. Conventional PCR (cPCR) tests, combined with Restriction Fragment Length Polymorphism (RFLP) or sequencing, were developed for FAdV diagnosis. Although these molecular tests have considerably improved the accuracy of FAdV diagnosis compared with conventional methods, certain drawbacks remain unresolved, including lack of sensitivity and post-PCR analysis. Subsequently, advanced molecular technologies such as real-time PCR (qPCR), Loop Isothermal Amplification (LAMP), Cross-Priming Amplification (CPA), Recombinase Polymerase Amplification (RPA), Digital Droplet Polymerase Chain Reaction (ddPCR), Dot Blot Assay Combined with cPCR, Nanoparticle-Assisted PCR (nano-PCR), PCR-Refractory Quantitative Amplification (ARMS-qPCR), CRISPR/Cas13a Technology, and High-Resolution Melting Curve (HRM), have been developed to improve FAdV diagnosis.
Keywords: CRISPR/Cas13; HRM; fowl adenovirus; genotyping; isothermal amplification; molecular diagnosis; polymerase chain reaction; real-time PCR.
Copyright © 2025 Kardoudi, Siham, Abdelmounaaim, Faouzi, Ikram, Thomas and Abdelouaheb.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Rapid detection of FAdV-4 by one-tube RPA-CRISPR/Cas12a assay.Front Microbiol. 2025 Feb 3;16:1541943. doi: 10.3389/fmicb.2025.1541943. eCollection 2025. Front Microbiol. 2025. PMID: 39963492 Free PMC article.
-
Application of cross-priming amplification (CPA) for detection of fowl adenovirus (FAdV) strains.Arch Virol. 2015 Apr;160(4):1005-13. doi: 10.1007/s00705-015-2355-9. Epub 2015 Feb 6. Arch Virol. 2015. PMID: 25655263 Free PMC article.
-
Development of a Multiplex Conventional PCR Assay for Concurrent Detection of FAdV-4, FAdV-8b, and FAdV-11.Vet Sci. 2025 Feb 17;12(2):177. doi: 10.3390/vetsci12020177. Vet Sci. 2025. PMID: 40005937 Free PMC article.
-
Fowl adenovirus-induced diseases and strategies for their control - a review on the current global situation.Avian Pathol. 2018 Apr;47(2):111-126. doi: 10.1080/03079457.2017.1385724. Epub 2017 Oct 25. Avian Pathol. 2018. PMID: 28950714 Review.
-
Fowl Adenovirus Serotype 1: From Gizzard Erosion to Comprehensive Insights into Genome Organization, Epidemiology, Pathogenesis, Diagnosis, and Prevention.Vet Sci. 2025 Apr 17;12(4):378. doi: 10.3390/vetsci12040378. Vet Sci. 2025. PMID: 40284880 Free PMC article. Review.
Cited by
-
The Detection and Differentiation of Pigeon Adenovirus Types 1 and 2 via a High-Resolution Melting Curve Platform.Microorganisms. 2025 Jun 7;13(6):1331. doi: 10.3390/microorganisms13061331. Microorganisms. 2025. PMID: 40572219 Free PMC article.
References
-
- Williams SM. A laboratory manual for the isolation, identification and characterization of avian pathogens. Jacksonville, FL: American Association of Avian Pathologists; (2016).
-
- International Committee on Taxonomy of Viruses. Van Regenmortel MHV, International Union of Microbiological Societies . Virus taxonomy: Classification and nomenclature of viruses: Seventh report of the international committee on taxonomy of viruses: Seventh report of the international committee on taxonomy of viruses. San Diego: Academic Press; (2000).
Publication types
LinkOut - more resources
Full Text Sources

