Identification of Hub Genes and Pathways in Preinfusion Chimeric Antigen Receptor (CAR) T-cell Products Associated With Cytokine Release Syndrome
- PMID: 40370902
- PMCID: PMC12076287
- DOI: 10.7759/cureus.82155
Identification of Hub Genes and Pathways in Preinfusion Chimeric Antigen Receptor (CAR) T-cell Products Associated With Cytokine Release Syndrome
Abstract
Background: Chimeric antigen receptor (CAR) T-cell therapy has transformed cancer management over the past decades, offering new hope to many patients. However, its effectiveness is often limited due to cytokine release syndrome (CRS), a life-threatening inflammatory response. Despite its clinical relevance, the molecular mechanisms underlying CRS, specifically in CAR T-cell products, remain poorly understood. This study aims to identify hub genes and pathways in preinfusion CAR T-cell products associated with CRS development and evaluate their potential as therapeutic targets through drug-gene interaction analysis and immune cell correlation profiling.
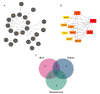
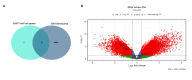
Methods: We examined gene expression data from 43 preinfusion clusters of differentiation 22 of CAR T-cell samples (CD22+), sourced from the Gene Expression Omnibus dataset GSE200296. Using the linear models for microarray data package, we identified differences in gene expression and conducted enrichment analyses to explore relevant biological pathways, including Kyoto Encyclopedia of Genes and Genomes and Gene Ontology terms. We built protein-protein interaction networks using the Search Tool for Retrieval of Interacting Genes/Proteins database to understand how these genes interact and pinpointed central "hub genes" with Cytoscape and the cytoHubba plugin. Our findings were validated using the GeneCards database (Weizmann Institute of Science, Israel) and an independent CRS-related dataset (GSE164805). Additionally, we analyzed immune cell populations and explored potential drug-gene interactions.
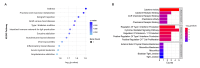
Results: Our study identified 24 genes with changed expression levels: 16 were downregulated and eight were upregulated. We identified five hub genes, interleukin (IL)1B, IL15, CD276, NCR2, and CCL17, as key contributors in CRS, which were primarily implicated in immune-related pathways, including cytokine-cytokine receptor interactions, IL17 signaling, and TNF signaling. These genes were especially expressed in monocytes, macrophages, and dendritic cells, confirming that those immune cell types play a critical role in CRS development. Through drug-gene interaction analysis, we found prospective therapies, such as enoblituzumab (targeting CD276) and canakinumab (targeting IL1B), which might assist in reducing CRS severity.
Conclusion: The study highlights IL1B, IL15, CD276, NCR2, and CCL17 as key CRS genes in preinfusion CAR T-cell products. Their dysregulation activity may contribute to the increased inflammation noted in CRS, pointing to a loss of regulatory control. Bringing us closer to better patient outcomes, these findings not only suggest that these genes could serve as valuable biomarkers for predicting CRS but also open the way for the development of more precise treatments such as combining drugs such as enoblituzumab and canakinumab, which might assist in reducing CRS severity and making CAR T-cell therapy safer and more effective, ultimately improving patient lives.
Keywords: bioinformatics analysis; car t-cell therapy; cytokine release syndrome; enrichment pathways analysis; hub genes.
Copyright © 2025, Khalafallah et al.
Conflict of interest statement
Human subjects: All authors have confirmed that this study did not involve human participants or tissue. Animal subjects: All authors have confirmed that this study did not involve animal subjects or tissue. Conflicts of interest: In compliance with the ICMJE uniform disclosure form, all authors declare the following: Payment/services info: All authors have declared that no financial support was received from any organization for the submitted work. Financial relationships: All authors have declared that they have no financial relationships at present or within the previous three years with any organizations that might have an interest in the submitted work. Other relationships: All authors have declared that there are no other relationships or activities that could appear to have influenced the submitted work.
Figures





Similar articles
-
Identification of Hub Genes and Pathways Associated with CAR-T Cell-Mediated Neurotoxicity in DLBCL.Clin Lab. 2021 Aug 1;67(8). doi: 10.7754/Clin.Lab.2020.201213. Clin Lab. 2021. PMID: 34383407
-
Granulocyte-macrophage colony-stimulating factor inactivation in CAR T-cells prevents monocyte-dependent release of key cytokine release syndrome mediators.J Biol Chem. 2019 Apr 5;294(14):5430-5437. doi: 10.1074/jbc.AC119.007558. Epub 2019 Feb 25. J Biol Chem. 2019. PMID: 30804212 Free PMC article.
-
A Computational Model of Cytokine Release Syndrome during CAR T-cell Therapy.Adv Ther (Weinh). 2022 Oct;5(10):2200130. doi: 10.1002/adtp.202200130. Epub 2022 Aug 4. Adv Ther (Weinh). 2022. PMID: 36590643 Free PMC article.
-
Tocilizumab for the treatment of chimeric antigen receptor T cell-induced cytokine release syndrome.Expert Rev Clin Immunol. 2019 Aug;15(8):813-822. doi: 10.1080/1744666X.2019.1629904. Epub 2019 Jun 20. Expert Rev Clin Immunol. 2019. PMID: 31219357 Free PMC article. Review.
-
Spotlight on Tocilizumab in the Treatment of CAR-T-Cell-Induced Cytokine Release Syndrome: Clinical Evidence to Date.Ther Clin Risk Manag. 2020 Aug 4;16:705-714. doi: 10.2147/TCRM.S223468. eCollection 2020. Ther Clin Risk Manag. 2020. PMID: 32801727 Free PMC article. Review.
References
-
- CAR T cell immunotherapy for human cancer. June CH, O'Connor RS, Kawalekar OU, Ghassemi S, Milone MC. Science. 2018;359:1361–1365. - PubMed
-
- Tisagenlecleucel in adult relapsed or refractory diffuse large B-cell lymphoma. Schuster SJ, Bishop MR, Tam CS, et al. N Engl J Med. 2019;380:45–56. - PubMed
-
- Cytokine release syndrome and neurotoxicity following CAR T-cell therapy for hematologic malignancies. Freyer CW, Porter DL. J Allergy Clin Immunol. 2020;146:940–948. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
