Loss-of-function mutations in the fruit softening gene POLYGALACTURONASE1 doubled fruit firmness in strawberry
- PMID: 40371060
- PMCID: PMC12077169
- DOI: 10.1093/hr/uhae315
Loss-of-function mutations in the fruit softening gene POLYGALACTURONASE1 doubled fruit firmness in strawberry
Abstract
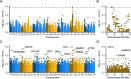
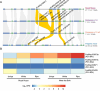
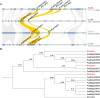
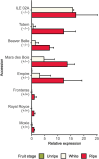
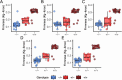
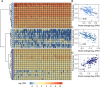
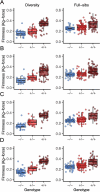
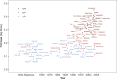
Wildtype fruit of cultivated strawberry (Fragaria [Formula: see text] ananassa) are typically soft and highly perishable when fully ripe. The development of firm-fruited cultivars by phenotypic selection has greatly increased shelf-life, decreased postharvest perishability, and driven the expansion of strawberry production worldwide. Hypotheses for the firm-fruited phenotype include mutations affecting the expression of genes encoding polygalacturonases (PGs) that soften fruit by degrading cell wall pectins. Here we show that loss-of-function mutations in the fruit softening gene POLYGALACTURONASE1 (FaPG1; PG1-6A1) double fruit firmness in strawberry. PG1-6A1 was one of three tandemly duplicated PG genes found to be in linkage disequilibrium (LD) with a quantitative trait locus (QTL) affecting fruit firmness on chromosome 6A. PG1-6A1 was strongly expressed in soft-fruited (wildtype) homozygotes and weakly expressed in firm-fruited (mutant) homozygotes. Genome-wide association, quantitative trait transcript, DNA sequence, and expression-QTL analyses identified genetic variants in LD with PG1-6A1 that were positively correlated with fruit firmness and negatively correlated with PG1-6A1 expression. An Enhancer/Suppressor-mutator (En/Spm) transposable element insertion was discovered upstream of PG1-6A1 in mutant homozygotes that we hypothesize transcriptionally downegulates the expression of PG1-6A1. The PG1-6A1 locus was incompletely dominant and explained 26-76% of the genetic variance for fruit firmness among phenotypically diverse individuals. Additional loci are hypothesized to underlie the missing heritability. Highly accurate codominant genotyping assays were developed for modifying fruit firmness by marker-assisted selection of the En/Spm insertion and single nucleotide polymorphisms associated with the PG1-6A1 locus.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nanjing Agricultural University.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Genome-wide association studies in a diverse strawberry collection unveil loci controlling agronomic and fruit quality traits.Plant Genome. 2024 Dec;17(4):e20509. doi: 10.1002/tpg2.20509. Epub 2024 Oct 15. Plant Genome. 2024. PMID: 39406253 Free PMC article.
-
QTL dynamics for fruit firmness and softening around an ethylene-dependent polygalacturonase gene in apple (Malus x domestica Borkh.).J Exp Bot. 2010 Jun;61(11):3029-39. doi: 10.1093/jxb/erq130. Epub 2010 May 12. J Exp Bot. 2010. PMID: 20462945 Free PMC article.
-
Down-regulation of POLYGALACTURONASE1 alters firmness, tensile strength and water loss in apple (Malus x domestica) fruit.BMC Plant Biol. 2012 Aug 2;12:129. doi: 10.1186/1471-2229-12-129. BMC Plant Biol. 2012. PMID: 22856470 Free PMC article.
-
Characterization of expansin genes and their transcriptional regulation by histone modifications in strawberry.Planta. 2021 Jul 3;254(2):21. doi: 10.1007/s00425-021-03665-6. Planta. 2021. PMID: 34216276 Review.
-
Molecular bases of strawberry fruit quality traits: Advances, challenges, and opportunities.Plant Physiol. 2023 Sep 22;193(2):900-914. doi: 10.1093/plphys/kiad376. Plant Physiol. 2023. PMID: 37399254 Review.
References
-
- Fletcher SW. Strawberry-Growing. New York: The Macmillan Company; 1917
-
- Darrow GM. The Strawberry: History, Breeding, and Physiology. New York: Holt, Rinehart, and Winston; 1966
-
- Lawrence F, Galletta G, Scott D. Strawberry breeding work of the US Department of Agriculture. HortScience. 1990;25:895–6
-
- Shaw DV, Larson KD. Performance of early-generation and modern strawberry cultivars from the University of California breeding programme in growing systems simulating traditional and modern horticulture. J Hortic Sci Biotechnol. 2008;83:648–52
Associated data
LinkOut - more resources
Full Text Sources
Research Materials

