Deletion Testing of the DEGS1 Gene Should Be Part of the Diagnostic Pipeline for Hypomyelinating Leukodystrophy (HLD18)
- PMID: 40371095
- PMCID: PMC12077970
- DOI: 10.1155/humu/3531508
Deletion Testing of the DEGS1 Gene Should Be Part of the Diagnostic Pipeline for Hypomyelinating Leukodystrophy (HLD18)
Abstract
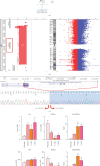
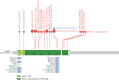
Hypomyelinating leukodystrophies are a heterogeneous group of disorders characterized by abnormal myelin formation in the central nervous system. Thanks to the increased use of NGS, a growing number of pathogenic single nucleotide variants in DEGS1 have recently been reported to be responsible for hypomyelinating leukodystrophy 18 (HLD18), a rare and severe autosomal recessive form. DEGS1 is a small gene (4 exons and 17 kb) encoding Δ4-dihydroceramide desaturase, which catalyzes the final step in ceramide biosynthesis. Here, we present two patients from unrelated families affected by severe and progressive white matter disease with developmental delay with or without regression and severe intellectual disability. Trio exome sequencing (ES) revealed in both probands two homozygous missense variants in the DEGS1 gene, p.Asp16His and p.Asn255Ser, both inherited from their heterozygous healthy mothers and with a noncarrier father. This curious finding of inconsistent segregation data raises the need for further testing. There is no MLPA test available for this gene, as no deletions have been reported. However, we tried a customized high-resolution 1 M CGH array, which was surprisingly positive in both cases: a 63-kb heterozygous deletion encompassing the entire gene in one proband and a 7-kb heterozygous deletion of Exons 2-3 in the second case. Previously reported cases of HLD18 have all been found to carry single nucleotide pathogenic variants in DEGS1, and the two patients described here are the first to carry whole or partial microdeletions involving DEGS1 that unmask pathogenic missense variants on the other allele. These two cases report the first examples of microdeletions of DEGS1 that unmask recessive allele pathogenic variants, underscoring the importance of considering whole or partial gene deletions in the diagnostic pipeline.
Copyright © 2025 Mariateresa Zanobio et al. Human Mutation published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

