Genomic characterization of norovirus and Sapovirus from a diarrhea outbreak in a school linked to heavy rainfall
- PMID: 40371105
- PMCID: PMC12075162
- DOI: 10.3389/fmicb.2025.1570161
Genomic characterization of norovirus and Sapovirus from a diarrhea outbreak in a school linked to heavy rainfall
Abstract
Background: Diarrhea poses a serious threat to human health, and rainfall is known to increase the incidence of diarrheal diseases. On July 7, 2024, a diarrhea outbreak occurred in a school in Sishui County, Jining City, Shandong Province, China, following heavy rainfall. This study aimed to identify the pathogens responsible for the outbreak and characterize their whole genomes.
Methods: On July 8, 2024, a total of 21 stool samples from diarrhea cases, 2 water samples from private wells, and 1 drinking water sample from the school cafeteria were collected. Real-time quantitative PCR was used to detect Rotavirus A (RV-A), Norovirus genogroup I (NV GI), Norovirus genogroup II (NV GII), Sapovirus (SaV), Human Astrovirus (HAstV), and Human Adenovirus (HAdV). Whole-genome sequencing was performed for NV GI and SaV-positive samples to determine their genotypes, construct phylogenetic trees, and analyze amino acid variation sites in encoded proteins.
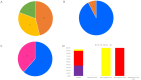
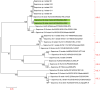
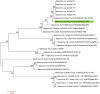
Results: Among the 21 case stool samples, 7 tested positive for both NV GI and SaV, 10 were positive for NV GI only, and 1 was positive for SaV only. Of the 2 private well water samples, one was positive for NV GI and the other for SaV. Whole-genome sequences were obtained for 11 NV GI strains and 2 SaV strains. The 11 NV GI sequences from the outbreak exhibited high homology, with whole-genome similarity ranging from 99.96% to 100%, and were all identified as the NV GI.6 [P11] genotype. Phylogenetic analysis showed that these 11 sequences clustered within the same evolutionary branch. Similarly, the 2 SaV sequences were highly homologous, with 99.97% similarity, and were identified as the SaV GI.6 genotype, clustering within the same phylogenetic branch.
Conclusions: This diarrhea outbreak was caused by the combined presence of NV GI and SaV following heavy rainfall. These findings provide valuable reference data for the prevention and control of diarrhea outbreaks caused by heavy rainfall or multiple pathogens.
Keywords: Norovirus; Sapovirus; diarrhea outbreak; heavy rainfall; whole genome.
Copyright © 2025 Fu, Wu, Yu, Zhao, Dong, Liu, Dou, Shi, Cai, Jiao, Liu and Jiao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Molecular epidemiology and characterization of norovirus and sapovirus in pediatric patients with acute diarrhea in Thailand, 2019-2020.J Infect Public Health. 2022 Sep;15(9):1013-1019. doi: 10.1016/j.jiph.2022.08.006. Epub 2022 Aug 15. J Infect Public Health. 2022. PMID: 35994998
-
Epidemiological Characteristics and Genotypic Features of Rotavirus and Norovirus in Jining City, 2021-2022.Viruses. 2024 Jun 7;16(6):925. doi: 10.3390/v16060925. Viruses. 2024. PMID: 38932216 Free PMC article.
-
Molecular Epidemiology of Human Adenovirus, Astrovirus, and Sapovirus Among Outpatient Children With Acute Diarrhea in Chongqing, China, 2017-2019.Front Pediatr. 2022 Mar 3;10:826600. doi: 10.3389/fped.2022.826600. eCollection 2022. Front Pediatr. 2022. PMID: 35311045 Free PMC article.
-
Molecular characterization of norovirus and sapovirus detected in animals and humans in Costa Rica: Zoo-anthropozoonotic potential of human norovirus GII.4.Open Vet J. 2023 Jan;13(1):74-89. doi: 10.5455/OVJ.2023.v13.i1.8. Epub 2023 Jan 14. Open Vet J. 2023. PMID: 36777439 Free PMC article.
-
Molecular epidemiology and genotype distributions of noroviruses and sapoviruses in Thailand 2000-2016: A review.J Med Virol. 2018 Apr;90(4):617-624. doi: 10.1002/jmv.25019. Epub 2018 Jan 23. J Med Virol. 2018. PMID: 29315631 Review.
References
-
- Cohen A. L., Platts-Mills J. A., Nakamura T., Operario D. J., Antoni S., Mwenda J. M., et al. . (2022). Aetiology and incidence of diarrhoea requiring hospitalisation in children under 5 years of age in 28 low-income and middle-income countries: findings from the Global Pediatric Diarrhea Surveillance network. BMJ Global Health 7:e009548. 10.1136/bmjgh-2022-009548 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

