Rapid extended-spectrum beta-lactamase-confirmation by using a machine learning model directly on routine automated susceptibility testing results
- PMID: 40371115
- PMCID: PMC12075368
- DOI: 10.3389/fmicb.2025.1582703
Rapid extended-spectrum beta-lactamase-confirmation by using a machine learning model directly on routine automated susceptibility testing results
Abstract
Objectives: Phenotypical Extended Spectrum β-Lactamase (ESBL)-production is commonly determined using the combination disk diffusion test or gradient test. This requires overnight incubation, prolonging time-to-detection and increasing duration of empirical treatment for patients with infections caused by gram-negative bacteria. To achieve instant confirmation without incubation, we developed a machine learning (ML)-model that predicts phenotypic ESBL-confirmation using Minimum Inhibitory Concentrations from routine automated antimicrobial susceptibility testing (AST)-results.
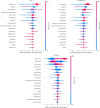
Methods: Data from the Dutch national laboratory-based surveillance system ISIS-AR collected between 2013 and 2022 from 49 laboratories were used: 178,044 isolates of E. coli (141,576), K. pneumoniae (33,088), and P. mirabilis (3,380) that exhibited resistance to cefotaxime and/or ceftazidime, and had available results of phenotypical ESBL-confirmation testing. We evaluated Logistic Regression, Random Forest and XGBoost models and calculated SHAP-values (SHapley Additive exPlanations) to identify most contributing features. We externally validated models using 5,996 isolates collected in Amsterdam University Medical Centres' between 2013 and 2022.
Results: XGBoost achieved an AUROC (Area Under Receiver Operating Characteristics) of 0.97, a sensitivity of 0.89 and an accuracy of 0.93. The most contributing features were the antibiotics cefotaxime, cefoxitin and trimethoprim for E. coli and K. pneumoniae, and cefuroxime, imipenem and cefotaxime for P. mirabilis. External validation yielded AUROCs of 0.93 (E. coli), 0.89 (K. pneumoniae) and 0.93 (P. mirabilis).
Conclusion: ML-models for prediction of ESBL-production using routine AST-system data achieved high performances. Implementing these models in laboratory practice could shorten time-to-detection. Once deployed, this approach could facilitate widespread screening for phenotypic ESBL-production.
Keywords: ESBL; antimicrobial resistance; bacteria; machine learning; surveillance.
Copyright © 2025 Ghouch, Schut, Sigaloff, Altorf-Van Der Kuil, Prins, Schade and the ISIS-AR study group.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Extended spectrum and metalo beta lactamase producing gram negative bacterial pathogens from cockroaches collected at hospital, Southern Ethiopia.Antimicrob Resist Infect Control. 2024 Aug 13;13(1):87. doi: 10.1186/s13756-024-01442-0. Antimicrob Resist Infect Control. 2024. PMID: 39135145 Free PMC article.
-
Antimicrobial resistance profile of extended-spectrum beta-lactamases, adenosine-monophosphate-cyclic, and carbapenemase-producing Gram-negative bacteria isolated from domestic animals.Vet World. 2021 Dec;14(12):3099-3104. doi: 10.14202/vetworld.2021.3099-3104. Epub 2021 Dec 13. Vet World. 2021. PMID: 35153398 Free PMC article.
-
Extended spectrum beta-lactamase and metallo beta-lactamase production among Escherichia coli and Klebsiella pneumoniae isolated from different clinical samples in a tertiary care hospital in Kathmandu, Nepal.Ann Clin Microbiol Antimicrob. 2017 Sep 19;16(1):62. doi: 10.1186/s12941-017-0236-7. Ann Clin Microbiol Antimicrob. 2017. PMID: 28927454 Free PMC article.
-
Evaluation of the Clinical and Laboratory Standards Institute phenotypic confirmatory test to detect the presence of extended-spectrum β-lactamases from 4005 Escherichia coli, Klebsiella oxytoca, Klebsiella pneumoniae and Proteus mirabilis isolates.J Med Microbiol. 2014 Apr;63(Pt 4):556-561. doi: 10.1099/jmm.0.068981-0. Epub 2014 Jan 29. J Med Microbiol. 2014. PMID: 24478449
-
Prevalence of Extended Spectrum β-lactamase-Producing Klebsiella pneumoniae in Clinical Isolates.Jundishapur J Microbiol. 2014 Nov;7(11):e17114. doi: 10.5812/jjm.17114. Epub 2014 Nov 1. Jundishapur J Microbiol. 2014. PMID: 25774279 Free PMC article.
References
-
- Abu-Aqil G., Suleiman M., Sharaha U., Nesher L., Lapidot I., Salman A., et al. (2023). Detection of extended-spectrum β-lactamase-producing bacteria isolated directly from urine by infrared spectroscopy and machine learning. Spectrochim. Acta A Mol. Biomol. Spectrosc. 295:122634. 10.1016/j.saa.2023.122634 - DOI - PubMed
-
- Altorf-van der Kuil W., Schoffelen A. F., de Greeff S. C., Thijsen S. F., Alblas H. J., Notermans D. W., et al. (2017). National laboratory-based surveillance system for antimicrobial resistance: A successful tool to support the control of antimicrobial resistance in the Netherlands. Euro Surveill. 22 17–00062. 10.2807/1560-7917.ES.2017.22.46.17-00062 - DOI - PMC - PubMed
-
- Boattini M., Bianco G., Comini S., Iannaccone M., Casale R., Cavallo R., et al. (2022). Direct detection of extended-spectrum-β-lactamase-producers in Enterobacterales from blood cultures: A comparative analysis. Eur. J. Clin. Microbiol. Infect. Dis. 41 407–413. 10.1007/s10096-021-04385-1 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

