Unleashing virus structural biology: Probing protein and membrane intermediates in the dynamic process of membrane fusion
- PMID: 40371173
- PMCID: PMC12075009
- DOI: 10.1017/qrd.2025.3
Unleashing virus structural biology: Probing protein and membrane intermediates in the dynamic process of membrane fusion
Abstract
Viruses are highly dynamic macromolecular assemblies. They undergo large-scale changes in structure and organization at nearly every stage of their infectious cycles from virion assembly to maturation, receptor docking, cell entry, uncoating and genome delivery. Understanding structural transformations and dynamics across the virus infectious cycle is an expansive area for research that that can also provide insight into mechanisms for blocking infection, replication, and transmission. Additionally, the processes viruses carry out serve as excellent model systems for analogous cellular processes, but in more accessible form. Capturing and analyzing these dynamic events poses a major challenge for many structural biological approaches due to the size and complexity of the assemblies and the heterogeneity and transience of the functional states that are populated. Here we examine the process of protein-mediated membrane fusion, which is carried out by specialized machinery on enveloped virus surfaces leading to delivery of the viral genome. Application of two complementary methods, cryo-electron tomography and structural mass spectrometry enable dynamic intermediate states in intact fusion systems to be imaged and probed, providing a new understanding of the mechanisms and machinery that drive this fundamental biological process.
Keywords: CryoEM structural biology; dynamics and function; integrative structural biology; membranes; virology.
© The Author(s) 2025.
Conflict of interest statement
The author declares that he has no competing financial interests.
Figures



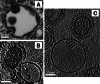

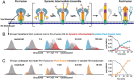
References
-
- Braet SM, Buckley TSC, Venkatakrishnan V, Dam KA, Bjorkman PJ and Anand GS (2023) Timeline of changes in spike conformational dynamics in emergent SARS-CoV-2 variants reveal progressive stabilization of trimer stalk with altered NTD dynamics. Elife 12, 1–21. 10.7554/eLife.82584. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
