Control of Physical and Biochemical Parameters Influencing Exogeneous Cargo Protein Association to Extracellular Vesicles Using Lipid Anchors Enables High Loading and Effective Intracellular Delivery
- PMID: 40371218
- PMCID: PMC12077271
- DOI: 10.1002/jex2.70048
Control of Physical and Biochemical Parameters Influencing Exogeneous Cargo Protein Association to Extracellular Vesicles Using Lipid Anchors Enables High Loading and Effective Intracellular Delivery
Abstract
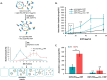
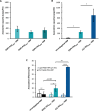
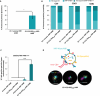
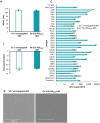
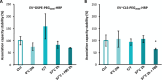
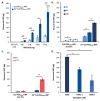
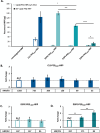
Despite biomolecule delivery is a natural function of extracellular vesicles (EVs), low loading of exogenous macromolecules such as proteins into EVs limits their interest as convincing protein delivery systems for health applications. In this context, lipid-anchorage of exogenous cargo into EV membrane recently emerged as a promising option to enable their vectorisation into cells. Nevertheless, this option was not explored for protein intracellular delivery, and further characterisation of critical parameters governing the association of a lipid-anchored cargo protein to EVs is still needed to confirm the relevance of this anchorage strategy. Therefore, we sought to identify these parameters in a precise and quantitative manner, using bulk and single nanoparticle analysis methods to identify protein loading capacity and subsequent intracellular delivery. We identified incubation temperature, cargo concentration, lipid anchor (LA) structure (lipid nature, linker) and EV origin as critical factors influencing maximal EV loading capacity. Precise control of these parameters enabled to load cargo protein close to EV saturation without hindering cellular delivery. The structural properties of LA influenced not only cargo protein/EV association but also intracellular delivery into different carcinoma cell lines. By thoroughly characterising Lipid-PEG-protein anchorage, this study evidences the interest of this tunable and controllable approach for efficient EV protein delivery.
Keywords: EV exogenous engineering; cell delivery; exosomes; lipid‐PEG anchorage; process optimisation; protein association.
© 2025 The Author(s). Journal of Extracellular Biology published by Wiley Periodicals LLC on behalf of International Society for Extracellular Vesicles.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
A platform for actively loading cargo RNA to elucidate limiting steps in EV-mediated delivery.J Extracell Vesicles. 2016 May 13;5:31027. doi: 10.3402/jev.v5.31027. eCollection 2016. J Extracell Vesicles. 2016. PMID: 27189348 Free PMC article.
-
Virus-Free Method to Control and Enhance Extracellular Vesicle Cargo Loading and Delivery.ACS Appl Bio Mater. 2023 Mar 20;6(3):1081-1091. doi: 10.1021/acsabm.2c00955. Epub 2023 Feb 13. ACS Appl Bio Mater. 2023. PMID: 36781171 Free PMC article.
-
Quantification of protein cargo loading into engineered extracellular vesicles at single-vesicle and single-molecule resolution.J Extracell Vesicles. 2021 Aug;10(10):e12130. doi: 10.1002/jev2.12130. Epub 2021 Aug 2. J Extracell Vesicles. 2021. PMID: 34377376 Free PMC article.
-
Breaking free: endocytosis and endosomal escape of extracellular vesicles.Extracell Vesicles Circ Nucl Acids. 2023 Jun 30;4(2):283-305. doi: 10.20517/evcna.2023.26. eCollection 2023. Extracell Vesicles Circ Nucl Acids. 2023. PMID: 39697985 Free PMC article. Review.
-
Current Strategies for Exosome Cargo Loading and Targeting Delivery.Cells. 2023 May 17;12(10):1416. doi: 10.3390/cells12101416. Cells. 2023. PMID: 37408250 Free PMC article. Review.
Cited by
-
Temporal and Severity-Dependent Alterations in Plasma Extracellular Vesicle Profiles Following Spinal Cord Injury.Cells. 2025 Jul 11;14(14):1065. doi: 10.3390/cells14141065. Cells. 2025. PMID: 40710318 Free PMC article.
References
-
- Andrews, M. D. , Dack K. N., de Groot M. J., et al. 2022. “Discovery of an Oral, Rule of 5 Compliant, Interleukin 17A Protein–Protein Interaction Modulator for the Potential Treatment of Psoriasis and Other Inflammatory Diseases.” Journal of Medicinal Chemistry 65: 8828–8842. - PubMed
-
- Choi, E. S. , Song J., Kang Y. Y., and Mok H.. 2019. “Mannose‐Modified Serum Exosomes for the Elevated Uptake to Murine Dendritic Cells and Lymphatic Accumulation.” Macromolecular Bioscience 19: 1900042. - PubMed
LinkOut - more resources
Full Text Sources
