A phylogenetic analysis of the CDKL protein family unravels its evolutionary history and supports the Drosophila model of CDKL5 deficiency disorder
- PMID: 40371392
- PMCID: PMC12075339
- DOI: 10.3389/fcell.2025.1582684
A phylogenetic analysis of the CDKL protein family unravels its evolutionary history and supports the Drosophila model of CDKL5 deficiency disorder
Abstract
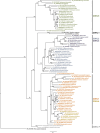
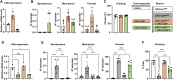
The human CDK-like (CDKL) family of serine‒threonine kinases has five members (CDKL1-5), with a conserved N-terminal kinase domain and variable C-termini. Among these, CDKL5 is of particular interest because of its involvement in CDKL5 deficiency disorder (CDD), a rare epileptic encephalopathy with several comorbidities for which there are no specific treatments. Current CDD vertebrate models are seizure resistant, which could be explained by the genetic background, including leaky expression of other CDKLs. Thus, phylogenetic analysis of the protein family would be valuable for understanding current models and developing new ones. Our phylogenetic studies revealed that ancestral CDKLs were present in all major eukaryotic clades and had ciliary/flagellar functions, which may have diversified throughout evolution. The original CDKL, which was likely similar to human CDKL5, gave rise to the remaining family members through successive duplications. In addition, particular clades have undergone further gene duplication and loss, a pattern that suggests some functional redundancy among them. A separate study focusing on the C-terminal tail of CDKL5 suggested that this domain is only functionally relevant in jawed vertebrates. We have developed a model of CDD in Drosophila based on downregulation of the single Cdkl gene by RNAi, which results in phenotypes similar to those of CDD patients, that are rescued by re-expression of fly Cdkl and human CDKL5. CDKL proteins contain a conserved kinase domain, originally involved in ciliary maintenance; therefore, invertebrate model organisms can be used to investigate CDKL functions that involve the aforementioned domain.
Keywords: CDKL5 deficiency disorder; ciliogenesis; cyclin-dependent-kinase like; epileptic encephalopathy; genetic redundancy; phylogeny; protein family.
Copyright © 2025 Martín-Carrascosa, Palacios-Martínez and Galindo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

