Solid stress compression enhances breast cancer cell migration through the upregulation of Interleukin-6
- PMID: 40371393
- PMCID: PMC12077316
- DOI: 10.3389/fcell.2025.1541953
Solid stress compression enhances breast cancer cell migration through the upregulation of Interleukin-6
Abstract
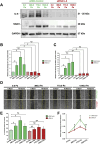
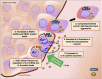
Apart from biochemical signals, tumour cells respond to biophysical and mechanical cues from their environment. The mechanical forces from the tumour microenvironment could be in the form of shear stress, tension, or solid stress compression. In this study, we explore the effects of solid stress compression on tumour cells. Solid stress compression, a prevalent biomechanical stimulus accumulated during tumour growth, has been shown to enhance invasive and metastatic phenotypes in cancer cells. However, the underlying molecular mechanism that elicits this aggressive metastatic phenotype, especially in breast cancer, is not extensively studied. Using an established 2D in vitro setup to apply incremental solid stress compression, we found that migratory and invasive capacities of aggressive breast cancer cells were enhanced in a biphasic manner. We also found that the transcript and protein levels of Interleukin-6 (IL-6) and SNAI1 were upregulated in response to solid stress. The resultant increased secretion of IL-6 could in turn lead to autocrine activation of downstream signalling pathways and impact on cancer cell migration and invasion.
Keywords: Interleukin-6; cell migration; mechanosensitive genes; solid stress compression; transcriptome.
Copyright © 2025 Azizan, Sheriff, Goh, Chiam and Koh.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
The matrix environmental and cell mechanical properties regulate cell migration and contribute to the invasive phenotype of cancer cells.Rep Prog Phys. 2019 Jun;82(6):064602. doi: 10.1088/1361-6633/ab1628. Epub 2019 Apr 4. Rep Prog Phys. 2019. PMID: 30947151 Review.
-
Compression enhances invasive phenotype and matrix degradation of breast Cancer cells via Piezo1 activation.BMC Mol Cell Biol. 2022 Jan 3;23(1):1. doi: 10.1186/s12860-021-00401-6. BMC Mol Cell Biol. 2022. PMID: 34979904 Free PMC article.
-
Interleukin-6/STAT3 signalling regulates adipocyte induced epithelial-mesenchymal transition in breast cancer cells.Sci Rep. 2018 Jun 11;8(1):8859. doi: 10.1038/s41598-018-27184-9. Sci Rep. 2018. PMID: 29891854 Free PMC article.
-
Senescence-associated IL-6 and IL-8 cytokines induce a self- and cross-reinforced senescence/inflammatory milieu strengthening tumorigenic capabilities in the MCF-7 breast cancer cell line.Cell Commun Signal. 2017 May 4;15(1):17. doi: 10.1186/s12964-017-0172-3. Cell Commun Signal. 2017. PMID: 28472950 Free PMC article.
-
The role and mechanism of compressive stress in tumor.Front Oncol. 2024 Sep 16;14:1459313. doi: 10.3389/fonc.2024.1459313. eCollection 2024. Front Oncol. 2024. PMID: 39351360 Free PMC article. Review.
References
-
- Blythe N. M., Muraki K., Ludlow M. J., Stylianidis V., Gilbert H. T. J., Evans E. L., et al. (2019). Mechanically activated Piezo1 channels of cardiac fibroblasts stimulate p38 mitogen-activated protein kinase activity and interleukin-6 secretion. J. Biol. Chem. 294, 17395–17408. 10.1074/jbc.RA119.009167 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

