Phylogenomics and functional annotation of 530 non- Saccharomyces yeasts from winemaking environments reveals their fermentome and flavorome
- PMID: 40371419
- PMCID: PMC12070155
- DOI: 10.3114/sim.2025.111.01
Phylogenomics and functional annotation of 530 non- Saccharomyces yeasts from winemaking environments reveals their fermentome and flavorome
Abstract
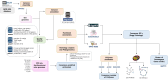
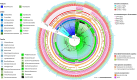
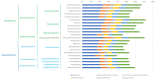
The winemaking industry faces unprecedented challenges due to climate change and market shifts, with profound commercial and socioeconomic repercussions. In response, non-Saccharomyces yeasts have gained attention for their potential to both mitigate these challenges and enhance the complexity of winemaking. This study builds upon our previous cataloguing of 293 non-Saccharomyces yeast species associated with winemaking environments by rigorously analysing 661 publicly available genomes. By employing a bioinformatics pipeline with stringent quality control checkpoints, we annotated and evaluated these genomes, culminating in a robust dataset of 530 non-Saccharomyces proteomes, belonging to 134 species, accessible to the research community. Employing this dataset, we conducted a comparative phylogenomic analysis to decipher metabolic networks related to fermentation capacity and flavor/aroma modulation. Our functional annotation has uncovered distinctive metabolic traits of non-Saccharomyces yeasts, elucidating their unique contributions to enology. Crucially, this work pioneers the identification of a non-Saccharomyces 'fermentome', a specific set of six genes uniquely present in fermentative species and absent in non-fermentative ones, and an expanded set of 35 genes constituting the complete fermentome. Moreover, we delineated a 'flavorome' by examining 96 genes across 19 metabolic categories implicated in wine aroma and flavour enhancement. These discoveries provide valuable genomic insights, offering new avenues for innovative winemaking practices and research. Citation: Franco-Duarte R, Fernandes T, Sousa MJ, Sampaio P, Rito T, Soares P (2025). Phylogenomics and functional annotation of 530 non-Saccharomyces yeasts from winemaking environments reveals their fermentome and flavorome. Studies in Mycology 111: 1-17. doi: 10.3114/sim.2025.111.01.
Keywords: bioinformatics; fermentation; fungi; genomics; non-conventional yeasts; phylogeny.
© 2025 Westerdijk Fungal Biodiversity Institute.
Figures


Similar articles
-
Beyond S. cerevisiae for winemaking: Fermentation-related trait diversity in the genus Saccharomyces.Food Microbiol. 2023 Aug;113:104270. doi: 10.1016/j.fm.2023.104270. Epub 2023 Mar 24. Food Microbiol. 2023. PMID: 37098430
-
Use of Non-Saccharomyces Yeasts in Berry Wine Production: Inspiration from Their Applications in Winemaking.J Agric Food Chem. 2022 Jan 26;70(3):736-750. doi: 10.1021/acs.jafc.1c07302. Epub 2022 Jan 12. J Agric Food Chem. 2022. PMID: 35019274 Review.
-
Selected non-Saccharomyces wine yeasts in controlled multistarter fermentations with Saccharomyces cerevisiae.Food Microbiol. 2011 Aug;28(5):873-82. doi: 10.1016/j.fm.2010.12.001. Epub 2010 Dec 10. Food Microbiol. 2011. PMID: 21569929
-
Interactions between Brettanomyces bruxellensis and other yeast species during the initial stages of winemaking.J Appl Microbiol. 2006 Jun;100(6):1208-19. doi: 10.1111/j.1365-2672.2006.02959.x. J Appl Microbiol. 2006. PMID: 16696668
-
Controlled mixed culture fermentation: a new perspective on the use of non-Saccharomyces yeasts in winemaking.FEMS Yeast Res. 2010 Mar;10(2):123-33. doi: 10.1111/j.1567-1364.2009.00579.x. Epub 2009 Sep 7. FEMS Yeast Res. 2010. PMID: 19807789 Review.
References
-
- Barata A, Malfeito-Ferreira M, Loureiro V. (2012). The microbial ecology of wine grape berries. International Journal of Food Microbiology 153: 243–259. - PubMed
Associated data
LinkOut - more resources
Full Text Sources
