A data-driven model for mitochondrial inner membrane remodeling as a driving force of organelle shaping
- PMID: 40371458
- PMCID: PMC12211560
- DOI: 10.1242/jcs.263850
A data-driven model for mitochondrial inner membrane remodeling as a driving force of organelle shaping
Abstract
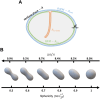
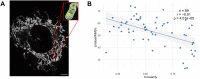
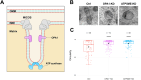
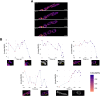
Mitochondria are dynamic organelles exhibiting diverse shapes. Although variation in mitochondrial shapes, which range from spheres to elongated tubules, and the transitions between them are clearly seen in many cell types, the molecular mechanisms governing this morphological variability remain poorly understood. Here, we propose a biophysical model for the shape transition between spheres and tubules based on the interplay between the inner and outer mitochondrial membranes. Our model suggests that the difference in surface area, arising from folding of the inner membrane into cristae, correlates with mitochondrial elongation. Analysis of live-cell super-resolution microscopy data supports this correlation, linking elongated shapes to the extent of cristae in the inner membrane. Knocking down cristae-shaping proteins further confirms the impact on mitochondrial shape, demonstrating that defects in cristae formation correlate with mitochondrial sphericity. Our results suggest that the dynamics of the inner mitochondrial membrane are not only important for simply creating surface area required for respiratory capacity but go beyond that to affect the whole organelle morphology. This work explores the biophysical foundations that govern the shape of individual mitochondria, suggesting potential links between mitochondrial structure and function. This should be of profound significance, particularly in the context of disrupted cristae-shaping proteins and their implications in mitochondrial diseases.
Keywords: Biophysical model; Cristae; Membrane remodeling; Mitochondrial membranes; Mitochondrial shape; Organelle shape.
© 2025. Published by The Company of Biologists.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures

Similar articles
-
Home treatment for mental health problems: a systematic review.Health Technol Assess. 2001;5(15):1-139. doi: 10.3310/hta5150. Health Technol Assess. 2001. PMID: 11532236
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Comparison of cellulose, modified cellulose and synthetic membranes in the haemodialysis of patients with end-stage renal disease.Cochrane Database Syst Rev. 2001;(3):CD003234. doi: 10.1002/14651858.CD003234. Cochrane Database Syst Rev. 2001. Update in: Cochrane Database Syst Rev. 2005 Jul 20;(3):CD003234. doi: 10.1002/14651858.CD003234.pub2. PMID: 11687058 Updated.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
-
Super-resolution microscopies, technological breakthrough to decipher mitochondrial structure and dynamic.Semin Cell Dev Biol. 2024 Jun-Jul;159-160:38-51. doi: 10.1016/j.semcdb.2024.01.006. Epub 2024 Feb 3. Semin Cell Dev Biol. 2024. PMID: 38310707 Review.
References
-
- Brakke, K. A. (1992). The surface evolver. Exp. Math. 1, 141-165. 10.1080/10586458.1992.10504253 - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

