IL-32-producing CD8+ memory T cells define immunoregulatory niches in human cutaneous leishmaniasis
- PMID: 40371647
- PMCID: PMC12077899
- DOI: 10.1172/JCI182040
IL-32-producing CD8+ memory T cells define immunoregulatory niches in human cutaneous leishmaniasis
Abstract
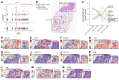
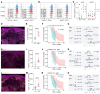
Human cutaneous leishmaniasis (CL) is characterized by chronic skin pathology. Experimental and clinical data suggest that immune checkpoints (ICs) play a crucial role in disease outcome, but the cellular and molecular niches that facilitate IC molecule expression during leishmaniasis are ill defined. In Sri Lankan patients with CL, indoleamine 2,3-dioxygenase 1 (IDO1) and programmed death-ligand 1 (PD-L1) were enriched in skin lesions, and reduced PD-L1 expression early after treatment initiation was predictive of a cure rate following antimonial therapy. Here, we used spatial cell interaction mapping to identify IL-32-expressing CD8+ memory T cells and Tregs as key components of the IDO1/PD-L1 niche in Sri Lankan patients with CL and in patients with distinct forms of dermal leishmaniasis in Brazil and India. Furthermore, the abundance of IL-32+ cells and IL-32+CD8+ T cells at treatment initiation was negatively correlated with the rate of cure in Sri Lankan patients. This study provides insights into the spatial mechanisms underpinning IC expression during CL and offers a strategy for identifying additional biomarkers of treatment response.
Keywords: Cellular immune response; Dermatology; Immunology; Infectious disease; Molecular pathology; Parasitology.
Conflict of interest statement
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

