Polyglutamine expansion induced dynamic misfolding of androgen receptor
- PMID: 40371721
- PMCID: PMC12079482
- DOI: 10.1002/pro.70154
Polyglutamine expansion induced dynamic misfolding of androgen receptor
Abstract
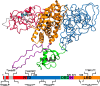
Spinal bulbar muscular atrophy (SBMA) is caused by a polyglutamine expansion (pQe) in the N-terminal transactivation domain of the human androgen receptor (AR-NTD), resulting in a combination of toxic gain- and loss-of-function mechanisms. The structural basis of these processes has not been resolved due to the disordered nature of the NTD, which hinders experimental analyses of its detailed conformations. Here, using extensive computational modeling, we show that AR-NTD forms dynamic compact regions, which upon pQe re-organize dynamically, mediated partly by direct pQ interaction with the Androgen N-Terminal Signature (ANTS) motif. The altered dynamics of the NTD result in a perturbation of interdomain interactions, with potential implications for the binding of the receptor protein to its response element. Oligomeric aggregation of the dynamic misfolded NTD exposes pQe, but blocks tau-5 and the FQNLF motif, which could lead to aberrant receptor transcriptional activity. These observations suggest a structural mechanism for AR dysfunction in SBMA.
Keywords: androgen receptor; intrinsically disordered protein; neuromuscular disorder; polyglutamine expansion; spinal‐bulbar muscular atrophy.
© 2025 The Author(s). Protein Science published by Wiley Periodicals LLC on behalf of The Protein Society.
Conflict of interest statement
The authors have no conflicts of interest to disclose.
Figures



References
-
- Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, et al. GROMACS: high performance molecular simulations through multi‐level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25.
-
- Adachi H. Widespread nuclear and cytoplasmic accumulation of mutant androgen receptor in SBMA patients. Brain. 2005;128:659–670. - PubMed
-
- Alibay I, Barnoud J, Beckstein O, Gowers RJ, Loche PR, MacDermott‐Opeskin H, et al. Building a community‐driven ecosystem for fast, reproducible, and reusable molecular simulation analysis using mdanalysis. Biophys J. 2023;122:420A.
-
- Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

