Hierarchy in regulator interactions with distant transcriptional activation domains empowers rheostatic regulation
- PMID: 40371733
- PMCID: PMC12079402
- DOI: 10.1002/pro.70142
Hierarchy in regulator interactions with distant transcriptional activation domains empowers rheostatic regulation
Abstract
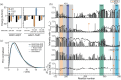
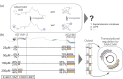
Transcription factors carry long intrinsically disordered regions often containing multiple activation domains. Despite numerous recent high-throughput identifications and characterizations of activation domains, the interplay between sequence motifs, activation domains, and regulator binding in intrinsically disordered transcription factor regions remains unresolved. Here, we map sequence motifs and activation domains in an Arabidopsis thaliana NAC transcription factor clade, revealing that although sequence motifs and activation domains often coincide, no systematic overlap exists. Biophysical analyses using NMR spectroscopy show that the long intrinsically disordered region of senescence-associated transcription factor ANAC046 is devoid of residual structure. We identify two activation domain/sequence motif regions, one at each end that both bind a panel of six positive and negative regulator domains from biologically relevant regulators promiscuously. Binding affinities measured using isothermal titration calorimetry reveal a hierarchy for regulator binding of the two ANAC046 activation domain/sequence motif regions defining these as regulatory hotspots. Despite extensive dynamic intramolecular contacts along the disordered chain revealed using paramagnetic relaxation enhancement experiments and simulations, the regions remain uncoupled in binding. Together, the results imply rheostatic regulation by ANAC046 through concentration-dependent regulator competition, a mechanism likely mirrored in other transcription factors with distantly located activation domains.
Keywords: activation domains; intrinsically disordered region; isothermal titration calorimetry; nuclear magnetic resonance; protein–protein interactions; short linear motif; transcription factor.
© 2025 The Author(s). Protein Science published by Wiley Periodicals LLC on behalf of The Protein Society.
Conflict of interest statement
KL‐L holds stock options and is a consultant for Peptone Ltd. All other authors declare no competing interests.
Figures



References
-
- Anderson JA, Glaser J, Glotzer SC. HOOMD‐blue: a Python package for high‐performance molecular dynamics and hard particle Monte Carlo simulations. Comput Mater Sci. 2020;173:109363.
-
- Bernadó P, Mylonas E, Petoukhov MV, Blackledge M, Svergun DI. Structural characterization of flexible proteins using small‐angle x‐ray scattering. J Am Chem Soc. 2007;129:5656–5664. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

