Benchmarking Zinc-Binding Site Predictors: A Comparative Analysis of Structure-Based Approaches
- PMID: 40371807
- PMCID: PMC12117554
- DOI: 10.1021/acs.jcim.5c00549
Benchmarking Zinc-Binding Site Predictors: A Comparative Analysis of Structure-Based Approaches
Erratum in
-
Correction to "Benchmarking Zinc-Binding Site Predictors: A Comparative Analysis of Structure-Based Approaches".J Chem Inf Model. 2025 Jul 14;65(13):7336. doi: 10.1021/acs.jcim.5c01366. Epub 2025 Jul 2. J Chem Inf Model. 2025. PMID: 40599056 Free PMC article. No abstract available.
Abstract
Metalloproteins play crucial physiological roles across all domains of life, relying on metal ions for structural stability and catalytic activity. In recent years, computational approaches have emerged as powerful and increasingly reliable tools for predicting metal-binding sites in metalloproteins, enabling their application in the challenging field of metalloproteomics. Given the growing number of available tools, it is timely to design a reproducible approach to characterize their performance in specific usage scenarios. Thus, in this study, we selected some state-of-the-art structure-based predictors for zinc-binding sites and evaluated their performance on two data sets: experimental apoprotein structures and structural models generated by AlphaFold. Our results indicate that apoprotein structures pose significant challenges for predicting metal-binding sites. For these systems, the predictors achieved lower-than-expected performance due to the structural rearrangements occurring upon metalation. Conversely, predictions based on AlphaFold models yielded significantly better results, suggesting that they more closely resemble the holo forms of metalloproteins. Our findings highlight the great potential of metal-binding site predictions for advancing research in the field of metalloproteomics.
Figures
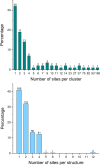

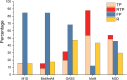


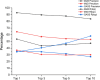


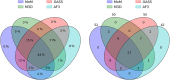

Similar articles
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
-
Interventions to reduce harm from continued tobacco use.Cochrane Database Syst Rev. 2016 Oct 13;10(10):CD005231. doi: 10.1002/14651858.CD005231.pub3. Cochrane Database Syst Rev. 2016. PMID: 27734465 Free PMC article.
-
Atypical antipsychotics for disruptive behaviour disorders in children and youths.Cochrane Database Syst Rev. 2017 Aug 9;8(8):CD008559. doi: 10.1002/14651858.CD008559.pub3. Cochrane Database Syst Rev. 2017. PMID: 28791693 Free PMC article.
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of paclitaxel, docetaxel, gemcitabine and vinorelbine in non-small-cell lung cancer.Health Technol Assess. 2001;5(32):1-195. doi: 10.3310/hta5320. Health Technol Assess. 2001. PMID: 12065068
References
-
- Jakubowski N., Lobinski R., Moens L.. Metallobiomolecules. The Basis of Life, the Challenge of Atomic Spectroscopy. J. Anal. At. Spectrom. 2004;19:1–4. doi: 10.1039/b313299b. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

