Unique molecular assay (UMA): a next-generation sequencing targeted panel for efficient and comprehensive genomic profiling and risk stratification of multiple myeloma
- PMID: 40371906
- PMCID: PMC12485320
- DOI: 10.3324/haematol.2025.287559
Unique molecular assay (UMA): a next-generation sequencing targeted panel for efficient and comprehensive genomic profiling and risk stratification of multiple myeloma
Abstract
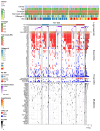
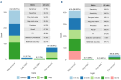
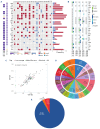
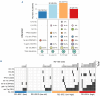
Multiple myeloma (MM) is characterized by genetic abnormalities in plasma cells, requiring precise genomic characterization for effective risk stratification and treatment. This study presents the Unique Molecular Assay (UMA) panel, a targeted DNA-sequencing approach designed to capture critical genomic aberrations in MM, including canonical immunoglobulin heavy chain translocations (t-IgH), copy number alterations (CNA), and mutations in 82 genes. The UMA panel is the first MM sequencing panel validated against traditional methods like fluorescence in situ hybridization (FISH) and SNP arrays across two laboratories for clinical-grade accuracy and reproducibility. The study included 150 patients whose DNA samples were analyzed using the UMA panel, achieving a median coverage of 233X with a requirement of ≥4 million reads per sample. The UMA panel demonstrated high concordance with FISH in detecting both CNA and t-IgH, achieving a balanced accuracy of over 93%. Moreover, inter-laboratory validation confirmed the robustness and reliability of the panel on genomic alteration calls. Importantly, the UMA panel enabled precise risk stratification based on the Second Revision of the International Staging System (R2-ISS), identifying high-risk features such as TP53 mutations and genome-wide CNA. This comprehensive and cost-effective genomic profiling tool supports clinical decision-making and personalized treatment strategies in MM. The validated performance and scalability of the UMA panel suggest its potential to complement traditional diagnostic methods, offering detailed insights into the genomic landscape of MM.
Figures


References
-
- Morgan GJ, Walker BA, Davies FE. The genetic architecture of multiple myeloma. Nat Rev Cancer. 2012;12(5):335-348. - PubMed
-
- Greipp PR, Miguel JS, Durie BGM, et al. International staging system for multiple myeloma. J Clin Oncol. 2005;23(15):3412-3420. - PubMed
-
- D’Agostino M, Cairns DA, Lahuerta JJ, et al. Second Revision of the International Staging System (R2-ISS) for overall survival in multiple myeloma: a European Myeloma Network (EMN) report within the HARMONY project. J Clin Oncol. 2022;40(29):3406-3418. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

