Tetrahymena ATG8 homologs, TtATG8A and TtATG8B, are responsible for mitochondrial degradation induced by starvation
- PMID: 40372018
- PMCID: PMC12153319
- DOI: 10.1128/mbio.00783-25
Tetrahymena ATG8 homologs, TtATG8A and TtATG8B, are responsible for mitochondrial degradation induced by starvation
Abstract
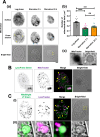
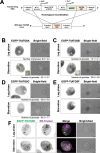
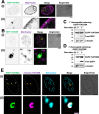
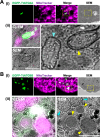
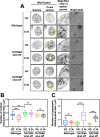
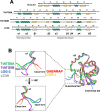
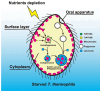
The majority of heterotrophic unicellular eukaryotes have evolved mechanisms to survive periods of starvation, allowing them to endure until conditions are favorable for regrowth. The ciliate Tetrahymena exhibits active swimming behavior in water, preying on microorganisms and growing exponentially at a rate of 0.5-0.75 h⁻¹ under optimal conditions. In this organism, numerous mitochondria localize to the cell cortex along the ciliary rows, likely ensuring an efficient ATP supply necessary for vigorous cell movement. Although mitochondrial reduction occurs immediately under starvation, the underlying mechanism remains unknown. Here, we demonstrated that autophagy is responsible for mitochondrial reduction in Tetrahymena thermophila. Among the five T. thermophila ATG8 homologs, TtATG8A and TtATG8B formed granule- and cup-shaped structures in response to starvation. Fluorescent microscopy further showed that TtATG8A and TtATG8B associate with mitochondria. Moreover, correlative light and electron microscopy analysis revealed that mitochondria colocalized with TtATG8A or TtATG8B were engulfed by autophagosomes and displayed abnormal appearances with disrupted cristae structures. Additionally, repression of TtATG8A or TtATG8B expression significantly attenuated starvation-induced mitochondrial reduction. These findings suggest that TtATG8A- and TtATG8B-mediated autophagy is a key mechanism underlying mitochondrial reduction in starved T. thermophila.
Importance: This study is the first comprehensive description of the mitochondrial degradation process under nutrient starvation in the ciliate Tetrahymena. It is well known that the cell surface structure of ciliates consists of an elaborate spatial arrangement of microtubule networks and associated structures and that this surface repetitive pattern is inherited by the next generation of cells like genetic information. Our findings provide a basis for understanding how ciliates maintain an adequate amount of mitochondria on the cell surface in response to nutritional conditions. Furthermore, we have successfully demonstrated the usefulness of Tetrahymena as an experimental system for studying mitochondrial quality control and turnover. Further studies of Tetrahymena will facilitate comparative studies among diverse biological systems on how eukaryotes other than opisthokonta (yeast, cultured cells, etc.) control their mitochondria.
Keywords: ATG8; Tetrahymena; autophagy; mitochondria.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Role of ATG8 and autophagy in programmed nuclear degradation in Tetrahymena thermophila.Eukaryot Cell. 2012 Apr;11(4):494-506. doi: 10.1128/EC.05296-11. Epub 2012 Feb 24. Eukaryot Cell. 2012. PMID: 22366125 Free PMC article.
-
Septins stabilize mitochondria in Tetrahymena thermophila.Eukaryot Cell. 2008 Aug;7(8):1373-86. doi: 10.1128/EC.00085-08. Epub 2008 Jun 27. Eukaryot Cell. 2008. PMID: 18586950 Free PMC article.
-
Case Study of the Response of N6-Methyladenine DNA Modification to Environmental Stressors in the Unicellular Eukaryote Tetrahymena thermophila.mSphere. 2021 Jun 30;6(3):e0120820. doi: 10.1128/mSphere.01208-20. Epub 2021 May 28. mSphere. 2021. PMID: 34047647 Free PMC article.
-
N6-methyladenine DNA modification in the unicellular eukaryotic organism Tetrahymena thermophila.Eur J Protistol. 2017 Apr;58:94-102. doi: 10.1016/j.ejop.2016.12.003. Epub 2016 Dec 23. Eur J Protistol. 2017. PMID: 28135687 Review.
-
Mapping the germ-line and somatic genomes of a ciliated protozoan, Tetrahymena thermophila.Genome Res. 1998 Feb;8(2):91-9. doi: 10.1101/gr.8.2.91. Genome Res. 1998. PMID: 9477337 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

