ASMC: investigating the amino acid diversity of enzyme active sites
- PMID: 40372409
- PMCID: PMC12133276
- DOI: 10.1093/bioinformatics/btaf307
ASMC: investigating the amino acid diversity of enzyme active sites
Abstract
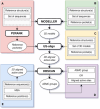
Motivation: The analysis of enzyme active sites is essential for understanding their activity in terms of catalyzed reaction and substrate specificity, providing insights for engineering to obtain targeted properties or modify the substrate scope. In 2010, a first version of the Active Site Modeling and Clustering (ASMC) workflow was published. ASMC predicts isofunctional clusters from enzyme families, based on structural modeling and clustering of active sites. Since then, structure- and sequence-based methods have developed considerably.
Results: We present here a redesign of the ASMC workflow. This new major version includes recent pocket prediction, structural alignment and clustering methods, as well as a refined amino acid distance matrix, thereby improving the relevance of results and reducing the need for laborious manual analysis to obtain relevant clusters. In addition, we have implemented multiple sequence alignment as a possible input for the clustering step, along with an additional script to compare 2D and 3D active sites. Finally, the code has been unified from three to one programming language (Python) to facilitate its installation and maintenance. This new version of ASMC was evaluated on a set of protein families, resulting in overall better performances compared to its original version.
Availability and implementation: ASMC is supported on Linux operating system and freely available at https://github.com/labgem/ASMC, along with a complete documentation (wiki, tutorial).
© The Author(s) 2025. Published by Oxford University Press.
Figures
Similar articles
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Factors that impact on the use of mechanical ventilation weaning protocols in critically ill adults and children: a qualitative evidence-synthesis.Cochrane Database Syst Rev. 2016 Oct 4;10(10):CD011812. doi: 10.1002/14651858.CD011812.pub2. Cochrane Database Syst Rev. 2016. PMID: 27699783 Free PMC article.
-
The clinical effectiveness and cost-effectiveness of enzyme replacement therapy for Gaucher's disease: a systematic review.Health Technol Assess. 2006 Jul;10(24):iii-iv, ix-136. doi: 10.3310/hta10240. Health Technol Assess. 2006. PMID: 16796930
References
-
- Bastard K, Perret A, Mariage A et al. Parallel evolution of non-homologous isofunctional enzymes in methionine biosynthesis. Nat Chem Biol 2017;13:858–66. - PubMed
-
- Bastard K, Smith AAT, Vergne-Vaxelaire C et al. Revealing the hidden functional diversity of an enzyme family. Nat Chem Biol 2014;10:42–9. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

