Shared host genetic landscape of respiratory viral infection
- PMID: 40372436
- PMCID: PMC12107129
- DOI: 10.1073/pnas.2414202122
Shared host genetic landscape of respiratory viral infection
Abstract
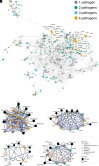
Respiratory viruses represent a major global health burden. Although these viruses have different life cycles, they may depend on common host genetic factors, which could be targeted by broad-spectrum host-directed therapies. We used genome-wide CRISPR screens and advanced data analytics to map a network of host genes that support infection by nine human respiratory viruses [influenza A virus, parainfluenza virus, human rhinovirus, respiratory syncytial virus, human coronavirus (HCoV)-229E, HCoV-NL63, HCoV-OC43, Middle East respiratory syndrome-related coronavirus, and severe acute respiratory syndrome-related coronavirus 2]. We explored shared pathways using knowledge graphs to inform on pharmacological targets. We selected and validated STT3A/B proteins of the N-oligosaccharyltransferase complex as host targets of broad-spectrum antiviral small molecules. Our work highlights the commonalities of viral host genetic dependencies and the feasibility of using this information to develop broad-spectrum antiviral agents.
Keywords: CRISPR; functional genomics; genome-wide; host-directed therapy; respiratory virus.
Conflict of interest statement
Competing interests statement:All authors were or are employees of Vir Biotechnology and may hold shares in Vir Biotechnology Inc. D.R.B., I.B., A.P., L.B.S., Z.N., W.T., J.G., S.H., A.T. have filed a patent related to this work.
Figures


References
-
- Ianevski A., et al. , DrugVirus.info 2.0: An integrative data portal for broad-spectrum antivirals (BSA) and BSA-containing drug combinations (BCCs). Nucleic Acids Res. 50, W272–W275 (2022). - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

