A Deep Learning-Enabled Workflow to Estimate Real-World Progression-Free Survival in Patients With Metastatic Breast Cancer: Study Using Deidentified Electronic Health Records
- PMID: 40372953
- PMCID: PMC12097284
- DOI: 10.2196/64697
A Deep Learning-Enabled Workflow to Estimate Real-World Progression-Free Survival in Patients With Metastatic Breast Cancer: Study Using Deidentified Electronic Health Records
Abstract
Background: Progression-free survival (PFS) is a crucial endpoint in cancer drug research. Clinician-confirmed cancer progression, namely real-world PFS (rwPFS) in unstructured text (ie, clinical notes), serves as a reasonable surrogate for real-world indicators in ascertaining progression endpoints. Response evaluation criteria in solid tumors (RECIST) is traditionally used in clinical trials using serial imaging evaluations but is impractical when working with real-world data. Manual abstraction of clinical progression from unstructured notes remains the gold standard. However, this process is a resource-intensive, time-consuming process. Natural language processing (NLP), a subdomain of machine learning, has shown promise in accelerating the extraction of tumor progression from real-world data in recent years.
objectives: We aim to configure a pretrained, general-purpose health care NLP framework to transform free-text clinical notes and radiology reports into structured progression events for studying rwPFS on metastatic breast cancer (mBC) cohorts.
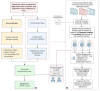
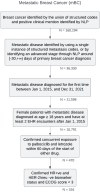
Methods: This study developed and validated a novel semiautomated workflow to estimate rwPFS in patients with mBC using deidentified electronic health record data from the Nference nSights platform. The developed workflow was validated in a cohort of 316 patients with hormone receptor-positive, human epidermal growth factor receptor-2 (HER-2) 2-negative mBC, who were started on palbociclib and letrozole combination therapy between January 2015 and December 2021. Ground-truth datasets were curated to evaluate the workflow's performance at both the sentence and patient levels. NLP-captured progression or a change in therapy line were considered outcome events, while death, loss to follow-up, and end of the study period were considered censoring events for rwPFS computation. Peak reduction and cumulative decline in Patient Health Questionnaire-8 (PHQ-8) scores were analyzed in the progressed and nonprogressed patient subgroups.
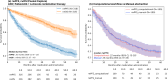
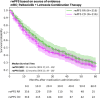
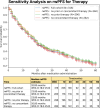
Results: The configured clinical NLP engine achieved a sentence-level progression capture accuracy of 98.2%. At the patient level, initial progression was captured within ±30 days with 88% accuracy. The median rwPFS for the study cohort (N=316) was 20 (95% CI 18-25) months. In a validation subset (n=100), rwPFS determined by manual curation was 25 (95% CI 15-35) months, closely aligning with the computational workflow's 22 (95% CI 15-35) months. A subanalysis revealed rwPFS estimates of 30 (95% CI 24-39) months from radiology reports and 23 (95% CI 19-28) months from clinical notes, highlighting the importance of integrating multiple note sources. External validation also demonstrated high accuracy (92.5% sentence level; 90.2% patient level). Sensitivity analysis revealed stable rwPFS estimates across varying levels of missing source data and event definitions. Peak reduction in PHQ-8 scores during the study period highlighted significant associations between patient-reported outcomes and disease progression.
Conclusions: This workflow enables rapid and reliable determination of rwPFS in patients with mBC receiving combination therapy. Further validation across more diverse external datasets and other cancer types is needed to ensure broader applicability and generalizability.
Keywords: EHR; ML; NLP; breast; cancer; data-driven oncology; deep learning; documentation; electronic health record; machine learning; metastatic; metastatic breast cancer; natural language processing; notes; oncology; real-world evidence; real-world progression-free survival; report; survival; workflow.
© Gowtham Varma, Rohit Kumar Yenukoti, Praveen Kumar M, Bandlamudi Sai Ashrit, K Purushotham, C Subash, Sunil Kumar Ravi, Verghese Kurien, Avinash Aman, Mithun Manoharan, Shashank Jaiswal, Akash Anand, Rakesh Barve, Viswanathan Thiagarajan, Patrick Lenehan, Scott A Soefje, Venky Soundararajan. Originally published in JMIR Cancer (https://cancer.jmir.org).
Conflict of interest statement
Figures
Similar articles
-
Comparative effectiveness of first-line palbociclib plus letrozole versus letrozole alone for HR+/HER2- metastatic breast cancer in US real-world clinical practice.Breast Cancer Res. 2021 Mar 24;23(1):37. doi: 10.1186/s13058-021-01409-8. Breast Cancer Res. 2021. PMID: 33761995 Free PMC article.
-
Machine learning and natural language processing (NLP) approach to predict early progression to first-line treatment in real-world hormone receptor-positive (HR+)/HER2-negative advanced breast cancer patients.Eur J Cancer. 2021 Feb;144:224-231. doi: 10.1016/j.ejca.2020.11.030. Epub 2020 Dec 26. Eur J Cancer. 2021. PMID: 33373867
-
Generating Real-World Tumor Burden Endpoints from Electronic Health Record Data: Comparison of RECIST, Radiology-Anchored, and Clinician-Anchored Approaches for Abstracting Real-World Progression in Non-Small Cell Lung Cancer.Adv Ther. 2019 Aug;36(8):2122-2136. doi: 10.1007/s12325-019-00970-1. Epub 2019 May 28. Adv Ther. 2019. PMID: 31140124 Free PMC article.
-
Systematic literature review of real-world evidence for treatments in HR+/HER2- second-line LABC/mBC after first-line treatment with CDK4/6i.BMC Cancer. 2024 May 23;24(1):631. doi: 10.1186/s12885-024-12269-8. BMC Cancer. 2024. PMID: 38783218 Free PMC article.
-
Leveraging Natural Language Processing and Machine Learning Methods for Adverse Drug Event Detection in Electronic Health/Medical Records: A Scoping Review.Drug Saf. 2025 Apr;48(4):321-337. doi: 10.1007/s40264-024-01505-6. Epub 2025 Jan 9. Drug Saf. 2025. PMID: 39786481 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

