Rev-RRE activity modulates HIV-1 replication and latency reactivation: Implications for viral persistence and cure strategies
- PMID: 40372991
- PMCID: PMC12080775
- DOI: 10.1371/journal.ppat.1012885
Rev-RRE activity modulates HIV-1 replication and latency reactivation: Implications for viral persistence and cure strategies
Abstract
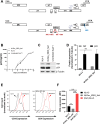
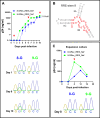
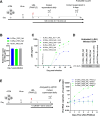
The HIV-1 Rev-RRE regulatory axis plays a crucial role in viral replication by facilitating the nucleo-cytoplasmic export and expression of viral mRNAs with retained introns. In this study, we investigated the impact of variation in Rev-RRE functional activity on HIV-1 replication kinetics and reactivation from latency. Using a novel HIV-1 viral vector with an interchangeable Rev cassette, we engineered viruses with two diverse Rev functional activities and demonstrated that higher Rev-RRE activity confers greater viral replication capacity while maintaining a constant level of Nef expression. In addition, a low Rev activity virus rapidly acquired a compensatory mutation in the RRE that significantly increased Rev-RRE activity and replication. In a latency model, proviruses with differing Rev-RRE activity levels varied in the efficiency of viral reactivation, affecting both initial viral release and subsequent replication kinetics. These results demonstrate that activity differences in the Rev-RRE axis among different viral isolates have important implications for HIV replication dynamics and persistence. Importantly, our findings indicate that bolstering Rev/RRE activity could be explored as part of latency reversal strategies in HIV cure efforts.
Copyright: © 2025 Dzhivhuho et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist
Figures


Update of
-
Rev-RRE activity modulates HIV-1 replication and latency reactivation: Implications for viral persistence and cure strategies.bioRxiv [Preprint]. 2025 Jan 6:2025.01.06.631466. doi: 10.1101/2025.01.06.631466. bioRxiv. 2025. Update in: PLoS Pathog. 2025 May 15;21(5):e1012885. doi: 10.1371/journal.ppat.1012885. PMID: 39829859 Free PMC article. Updated. Preprint.
References
-
- Bray M, Prasad S, Dubay JW, Hunter E, Jeang KT, Rekosh D, et al.. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc Natl Acad Sci U S A. 1994;91(4):1256–60. doi: 10.1073/pnas.91.4.1256 - DOI - PMC - PubMed
-
- Hadzopoulou-Cladaras M, Felber BK, Cladaras C, Athanassopoulos A, Tse A, Pavlakis GN. The rev (trs/art) protein of human immunodeficiency virus type 1 affects viral mRNA and protein expression via a cis-acting sequence in the env region. J Virol. 1989;63(3):1265–74. doi: 10.1128/JVI.63.3.1265-1274.1989 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

