Genomic insights into the serovar prevalence, antimicrobial resistance gene, and genetic diversity of Salmonella enterica in Mexico
- PMID: 40373083
- PMCID: PMC12080759
- DOI: 10.1371/journal.pone.0323872
Genomic insights into the serovar prevalence, antimicrobial resistance gene, and genetic diversity of Salmonella enterica in Mexico
Abstract
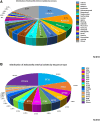
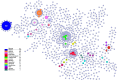
The research aims to provide insights into the sources of contamination, prevalence of common serovars, determination of sequence types, prediction of genes associated with antimicrobial resistance, and phylogenetic analysis to evaluate genetic diversity and correlations between serovars and sequence types in Salmonella enterica in Mexico. We analyzed 818 publicly accessible whole-genome sequences from Mexico, which included isolates from diverse sources such as poultry, meat, water, and agricultural environments. We identified fifty-seven serovars, of which 19.8% corresponded to S. Infantis, 10.7% to S. Anatum, and 6.6% to S. Newport, representing the most common serovars. Phylogenetic analysis shows a strong correlation between sequence type and serovar. For example, ST32 for S. Infantis and ST64 for S. Anatum show this. However, serovars such as S. Newport possessed considerable genomic diversity, suggesting complex contamination pathways. The analysis showed that many isolates have been identified as multidrug-resistant, exhibiting resistance gene profiles for aminoglycosides, β-lactams, fluoroquinolones, sulfonamides, and tetracyclines. The findings emphasize the importance of identifying contamination sources to monitor the dissemination of multidrug-resistant Salmonella in regions that have significant antibiotic consumption in agriculture and farming, highlighting its global relevance for food safety and public health.
Copyright: © 2025 Lozano-Aguirre et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Unveiling the genomic landscape of Salmonella enterica serotypes Typhimurium, Newport, and Infantis in Latin American surface waters: a comparative analysis.Microbiol Spectr. 2024 May 2;12(5):e0004724. doi: 10.1128/spectrum.00047-24. Epub 2024 Mar 28. Microbiol Spectr. 2024. PMID: 38546218 Free PMC article.
-
Genomic Epidemiology of Multidrug-Resistant Nontyphoidal Salmonella in Young Children Hospitalized for Gastroenteritis.Microbiol Spectr. 2021 Sep 3;9(1):e0024821. doi: 10.1128/Spectrum.00248-21. Epub 2021 Aug 4. Microbiol Spectr. 2021. PMID: 34346743 Free PMC article.
-
Whole-genome sequencing analysis and CRISPR genotyping of rare antibiotic-resistant Salmonella enterica serovars isolated from food and related sources.Food Microbiol. 2021 Feb;93:103601. doi: 10.1016/j.fm.2020.103601. Epub 2020 Aug 4. Food Microbiol. 2021. PMID: 32912589
-
High prevalence and genomic features of multidrug-resistant Salmonella enterica isolated from chilled broiler chicken on retail sale in the United Arab Emirates.Int J Food Microbiol. 2024 Oct 2;423:110828. doi: 10.1016/j.ijfoodmicro.2024.110828. Epub 2024 Jul 16. Int J Food Microbiol. 2024. PMID: 39032201
-
Genomic analysis of the MLST population structure and antimicrobial resistance genes associated with Salmonella enterica in Mexico.Genome. 2023 Dec 1;66(12):319-332. doi: 10.1139/gen-2023-0007. Epub 2023 Jul 21. Genome. 2023. PMID: 37478495
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

