High-resolution CTCF footprinting reveals impact of chromatin state on cohesin extrusion
- PMID: 40374602
- PMCID: PMC12081859
- DOI: 10.1038/s41467-025-57775-w
High-resolution CTCF footprinting reveals impact of chromatin state on cohesin extrusion
Abstract
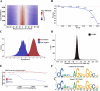
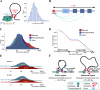
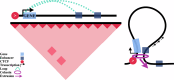
Cohesin-mediated DNA loop extrusion enables gene regulation by distal enhancers through the establishment of chromosome structure and long-range enhancer-promoter interactions. The best characterized cohesin-related structures, such as topologically associating domains (TADs) anchored at convergent CTCF binding sites, represent static conformations. Consequently, loop extrusion dynamics remain poorly understood. To better characterize static and dynamically extruding chromatin loop structures, we use MNase-based 3D genome assays to simultaneously determine CTCF and cohesin localization as well as the 3D contacts they mediate. Here we present CTCF Analyzer (with) Multinomial Estimation (CAMEL), a tool that identifies CTCF footprints at near base-pair resolution in CTCF MNase HiChiP. We also use Region Capture Micro-C to identify a CTCF-adjacent footprint that is attributed to cohesin occupancy. We leverage this substantial advance in resolution to determine that the fully extruded (CTCF-CTCF loop) state is rare genome-wide with locus-specific variation from ~1-10%. We further investigate the impact of chromatin state on loop extrusion dynamics and find that active regulatory elements impede cohesin extrusion. These findings support a model of topological regulation whereby the transient, partially extruded state facilitates enhancer-promoter contacts that can regulate transcription.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: Dovetail Genomics/Cantata Bio provided reagents and sample processing for HiChIP experiments. M.B. and M.S.B were employees at Dovetail Genomics during the course of this research. M.J.A has financial and consulting interests unrelated to this work in SeQure Dx and Chroma Medicine. M.J.A’s interests are reviewed and managed by Dana Farber Cancer Institute. J.K.J. is a co-founder of and has a financial interest in SeQure, Dx, Inc., a company developing technologies for gene editing target profiling. JKJ also has, or had during the course of this research, financial interests in several companies developing gene editing technology: Beam Therapeutics, Blink Therapeutics, Chroma Medicine, Editas Medicine, EpiLogic Therapeutics, Excelsior Genomics, Hera Biolabs, Monitor Biotechnologies, Nvelop Therapeutics (f/k/a ETx, Inc.), Pairwise Plants, Poseida Therapeutics, and Verve Therapeutics. J.K.J.’s interests were reviewed by Massachusetts General Hospital and Mass General Brigham in accordance with their conflict of interest policies. M.J.A., Y.E.T., and J.K.J. are currently employees of Arena Bioworks. The remaining authors declare no competing interests.
Figures



Update of
-
High-resolution CTCF footprinting reveals impact of chromatin state on cohesin extrusion dynamics.bioRxiv [Preprint]. 2023 Oct 28:2023.10.20.563340. doi: 10.1101/2023.10.20.563340. bioRxiv. 2023. Update in: Nat Commun. 2025 May 15;16(1):4506. doi: 10.1038/s41467-025-57775-w. PMID: 37961446 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
- NNF21SA0072102/Novo Nordisk Fonden (Novo Nordisk Foundation)
- T32GM135117/Foundation for the National Institutes of Health (Foundation for the National Institutes of Health, Inc.)
- T32 GM135117/GM/NIGMS NIH HHS/United States
- R35 GM118158/GM/NIGMS NIH HHS/United States
- RM1 HG009490/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources

