Tick-borne flavivirus exoribonuclease-resistant RNAs contain a double loop structure
- PMID: 40374626
- PMCID: PMC12081666
- DOI: 10.1038/s41467-025-59657-7
Tick-borne flavivirus exoribonuclease-resistant RNAs contain a double loop structure
Abstract
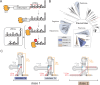
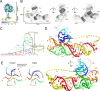
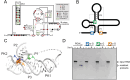
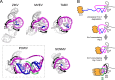
Viruses from the Flaviviridae family contain human relevant pathogens that generate subgenomic noncoding RNAs during infection using structured exoribonuclease resistant RNAs (xrRNAs). These xrRNAs block progression of host cell's 5' to 3' exoribonucleases. The structures of several xrRNAs from mosquito-borne and insect-specific flaviviruses reveal a conserved fold in which a ring-like motif encircles the 5' end of the xrRNA. However, the xrRNAs found in tick-borne and no known vector flaviviruses have distinct characteristics, and their 3-D fold was unsolved. Here, we verify the presence of xrRNAs in the encephalitis-causing tick-borne Powassan Virus. We characterize their secondary structure and obtain a mid-resolution map of one of these xrRNAs using cryo-EM, revealing a unique double-loop ring element. Integrating these results with covariation analysis, biochemical data, and existing high-resolution structural information yields a model in which the core of the fold matches the previously solved xrRNA fold, but the expanded double loop ring is remodeled upon encountering the exoribonuclease. These results are representative of a broad class of xrRNAs and reveal a conserved strategy of structure-based exoribonuclease resistance achieved through a unique topology across a viral family of importance to global health.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors have no competing interests.
Figures

References
-
- Mackenzie, J. S., Gubler, D. J. & Petersen, L. R. Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat. Med.10, S98–S109 (2004). - PubMed
-
- Valderrama, A., Diaz, Y. & Lopez-Verges, S. Interaction of Flavivirus with their mosquito vectors and their impact on the human health in the Americas. Biochem. Biophys. Res Commun.492, 541–547 (2017). - PubMed
MeSH terms
Substances
Grants and funding
- R01AI133348/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- U24GM129547/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- T32AI052066/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- F31AI156992/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- grid.436923.9/U.S. Department of Energy (DOE)
LinkOut - more resources
Full Text Sources

