GzESTY as an optimized cell-based assay for initial steps in GPCR deorphanization
- PMID: 40374633
- PMCID: PMC12081699
- DOI: 10.1038/s41467-025-59850-8
GzESTY as an optimized cell-based assay for initial steps in GPCR deorphanization
Abstract
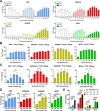
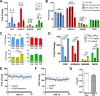
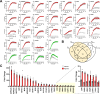
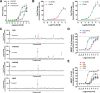
G protein-coupled receptors (GPCRs) are key pharmacological targets, yet many remain underutilized due to unknown activation mechanisms and ligands. Orphan GPCRs, lacking identified natural ligands, are a high priority for research, as identifying their ligands will aid in understanding their functions and potential as drug targets. Most GPCRs, including orphans, couple to Gi/o/z family members, however current assays to detect their activation are limited, hindering ligand identification efforts. We introduce GzESTY, a sensitive, cell-based assay developed in an easily deliverable format designed to study the pharmacology of Gi/o/z-coupled GPCRs and assist in deorphanization. We optimized assay conditions and developed an all-in-one vector employing cloning methods to ensure the correct expression ratio of GzESTY components. GzESTY successfully assessed activation of a library of ligand-activated GPCRs, detecting both full and partial agonism, and responses from endogenous GPCRs. Notably, with GzESTY we established the presence of endogenous ligands for GPR176 and GPR37 in brain extracts, validating its use in deorphanization efforts. This assay enhances the ability to find ligands for orphan GPCRs, expanding the toolkit for GPCR pharmacologists.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



Similar articles
-
Gz Enhanced Signal Transduction assaY (GZESTY) for GPCR deorphanization.bioRxiv [Preprint]. 2024 Jul 26:2024.07.26.605282. doi: 10.1101/2024.07.26.605282. bioRxiv. 2024. PMID: 39091869 Free PMC article. Preprint.
-
Deciphering complexity of GPCR signaling and modulation: implications and perspectives for drug discovery.Clin Sci (Lond). 2025 May 20;139(10):463-77. doi: 10.1042/CS20245182. Clin Sci (Lond). 2025. PMID: 40400289 Free PMC article. Review.
-
The Impact of Nanobodies on G Protein-Coupled Receptor Structural Biology and Their Potential as Therapeutic Agents.Mol Pharmacol. 2024 Sep 17;106(4):155-163. doi: 10.1124/molpharm.124.000974. Mol Pharmacol. 2024. PMID: 39107078 Review.
-
Fine-Tuned Deep Transfer Learning Models for Large Screenings of Safer Drugs Targeting Class A GPCRs.Biochemistry. 2025 Mar 18;64(6):1328-1337. doi: 10.1021/acs.biochem.4c00832. Epub 2025 Mar 8. Biochemistry. 2025. PMID: 40056143
-
Rapid molecular tests for tuberculosis and tuberculosis drug resistance: a qualitative evidence synthesis of recipient and provider views.Cochrane Database Syst Rev. 2022 Apr 26;4(4):CD014877. doi: 10.1002/14651858.CD014877.pub2. Cochrane Database Syst Rev. 2022. PMID: 35470432 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

