Towards global reaction feasibility and robustness prediction with high throughput data and bayesian deep learning
- PMID: 40374636
- PMCID: PMC12081921
- DOI: 10.1038/s41467-025-59812-0
Towards global reaction feasibility and robustness prediction with high throughput data and bayesian deep learning
Abstract
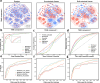
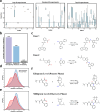
Predicting organic reaction feasibility and robustness against environmental factors is challenging. We address this issue by integrating high throughput experimentation (HTE) and Bayesian deep learning. Diverging from existing HTE studies focused on niche chemical spaces, in this work, our in-house HTE platform conducted 11,669 distinct acid amine coupling reactions in 156 working hours, yielding the most extensive single HTE dataset at a volumetric scale for industrial delivery. Our Bayesian neural network model achieved a benchmark for prediction accuracy of 89.48% for reaction feasibility. Furthermore, our fine-grained uncertainty disentanglement enables efficient active learning, reducing 80% of data requirements. Additionally, our uncertainty analysis effectively identifies out-of-domain reactions and evaluates reaction robustness or reproducibility against environmental factors for scaling up, offering a practical framework for navigating chemical spaces and designing highly robust industrial processes.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: H.Z., Y.L.L., H.S., Y.R.L., R.Z., B.L., Y.Y., S.L., P.W., and X.W. are employed by ChemLex, a company specializing in high-throughput synthesis. Y.H. serves as the Chief Executive Officer of MegaRobo, which provides high-throughput experimentation (HTE) equipment. The remaining authors (K.F., F.M., F.Y., and T.Y.) declare no competing interests.
Figures



Similar articles
-
Probing the chemical 'reactome' with high-throughput experimentation data.Nat Chem. 2024 Apr;16(4):633-643. doi: 10.1038/s41557-023-01393-w. Epub 2024 Jan 2. Nat Chem. 2024. PMID: 38168924 Free PMC article.
-
Ultrahigh-Throughput Experimentation for Information-Rich Chemical Synthesis.Acc Chem Res. 2021 May 18;54(10):2337-2346. doi: 10.1021/acs.accounts.1c00119. Epub 2021 Apr 23. Acc Chem Res. 2021. PMID: 33891404
-
The Evolution of Chemical High-Throughput Experimentation To Address Challenging Problems in Pharmaceutical Synthesis.Acc Chem Res. 2017 Dec 19;50(12):2976-2985. doi: 10.1021/acs.accounts.7b00428. Epub 2017 Nov 27. Acc Chem Res. 2017. PMID: 29172435
-
The challenge of balancing model sensitivity and robustness in predicting yields: a benchmarking study of amide coupling reactions.Chem Sci. 2023 Sep 13;14(39):10835-10846. doi: 10.1039/d3sc03902a. eCollection 2023 Oct 11. Chem Sci. 2023. PMID: 37829036 Free PMC article.
-
Developing Software for Using Bayesian Regression to Evaluate Heterogeneity of Treatment Effects in Data from Randomized Controlled Trials [Internet].Washington (DC): Patient-Centered Outcomes Research Institute (PCORI); 2018 Jul. Washington (DC): Patient-Centered Outcomes Research Institute (PCORI); 2018 Jul. PMID: 38620392 Free Books & Documents. Review.
References
-
- Sen, M., Arguelles, A. J., Stamatis, S. D., García-Muñoz, S. & Kolis, S. An optimization-based model discrimination framework for selecting an appropriate reaction kinetic model structure during early phase pharmaceutical process development. React. Chem. Eng.6, 2092–2103 (2021).
-
- Schwaller, P., Vaucher, A. C., Laino, T. & Reymond, J.-L. Prediction of chemical reaction yields using deep learning. Mach. Learn. Sci. Technol.2, 015016 (2021).
-
- Schleinitz, J. et al. Machine learning yield prediction from nicolit, a small-size literature data set of nickel catalyzed c–o couplings. J. Am. Chem. Soc.144, 14722–14730 (2022). - PubMed
LinkOut - more resources
Full Text Sources

