Tumor budding and poorly differentiated clusters as a biological continuum in colorectal cancer invasion and prognosis
- PMID: 40374662
- PMCID: PMC12081641
- DOI: 10.1038/s41598-025-00866-x
Tumor budding and poorly differentiated clusters as a biological continuum in colorectal cancer invasion and prognosis
Abstract
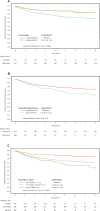
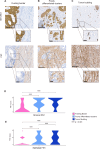
Tumor budding (TB) and poorly differentiated clusters (PDCs) are features of infiltrative growth patterns and powerful independent prognostic factors in colorectal cancer (CRC), yet the underlying biological mechanisms behind their role in CRC invasion is less understood. The aim of this study was to investigate the molecular background and prognostic role of tumor cluster size at the invasive margin (IM) of CRC, and determine whether a biological continuum between TB and PDCs exists. Using a combination of spatial transcriptomic and immunohistochemical (IHC) techniques, we demonstrated a biological continuum from larger to smaller tumor clusters, with TB possessing greater invasive potential than PDCs. We deployed artificial intelligence on a cohort of 1134 Stage I-III CRC resections to automatically detect nearly 400,000 isolated tumor cells/clusters of any particular size across the IM. We determined that 2-celled clusters were the most abundant feature at the IM, and the simultaneous assessment of TB and PDCs yielded a prognostic performance stronger than either independently. Our study provides a deeper understanding of the mechanisms behind CRC invasion while improving risk stratification for Stage I-III CRC.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: JvdL was a member of the advisory boards of Philips, the Netherlands and ContextVision, Sweden, and received research funding from Philips, the Netherlands, ContextVision, Sweden, and Sectra, Sweden in the last five years. He is chief scientific officer (CSO) and shareholder of Aiosyn BV, the Netherlands. FC was Chair of the Scientific and Medical Advisory Board of TRIBVN Healthcare, France, and received advisory board fees from TRIBVN Healthcare, France in the last five years. He is shareholder of Aiosyn BV, the Netherlands. All other authors declare no conflict of interest.
Figures




References
-
- Jass, J. R., Love, S. B. & Northover, J. M. A. A new prognostic classification of rectal cancer. Lancet329, 1303–1306 (1987). - PubMed
-
- Karamitopoulou, E. et al. Tumour border configuration in colorectal cancer: proposal for an alternative scoring system based on the percentage of infiltrating margin. Histopathology67, 464–473 (2015). - PubMed
-
- Lugli, A. et al. Recommendations for reporting tumor budding in colorectal cancer based on the international tumor budding consensus conference (ITBCC) 2016. Mod. Pathol.30, 1299–1311 (2017). - PubMed
-
- Lugli, A., Zlobec, I., Berger, M. D., Kirsch, R. & Nagtegaal, I. D. Tumour budding in solid cancers. Nat. Reviews Clin. Oncol.18, 101–115 (2020). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

