A self-assembled protein β-helix as a self-contained biofunctional motif
- PMID: 40374664
- PMCID: PMC12081914
- DOI: 10.1038/s41467-025-59873-1
A self-assembled protein β-helix as a self-contained biofunctional motif
Abstract
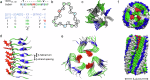
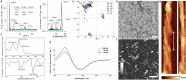
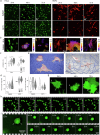
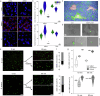
Nature constructs matter by employing protein folding motifs, many of which have been synthetically reconstituted to exploit function. A less understood motif whose structure-function relationships remain unexploited is formed by parallel β-strands arranged in a helical repetitive pattern, termed a β-helix. Herein we reconstitute a protein β-helix by design and endow it with biological function. Unlike β-helical proteins, which are contiguous covalent structures, this β-helix self-assembles from an elementary sequence of 18 amino acids. Using a combination of experimental and computational methods, we demonstrate that the resulting assemblies are discrete cylindrical structures exhibiting conserved dimensions at the nanoscale. We provide evidence for the structures to form a carpet-like three-dimensional scaffold promoting and inhibiting the growth of human and bacterial cells, respectively, while being able to mediate intracellular gene delivery. The study introduces a self-assembled β-helix as a self-contained bio- and multi-functional motif for exploring and exploiting mechanistic biology.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: B.W.H. holds an executive position at an AFM manufacturer Nanosurf. Nanosurf played no role in the design and execution of this study. B.W.H. has no other competing interests. The authors declare no other competing interests.
Figures

Similar articles
-
Structural motifs in which β-strands are clipped together with the П-like module.Proteins. 2017 Oct;85(10):1925-1930. doi: 10.1002/prot.25346. Epub 2017 Jul 16. Proteins. 2017. PMID: 28677205
-
Amino acid content of beta strands and alpha helices depends on their flanking secondary structure elements.Biosystems. 2018 Jun;168:45-54. doi: 10.1016/j.biosystems.2018.04.002. Epub 2018 May 6. Biosystems. 2018. PMID: 29742459
-
Salt-bridging effects on short amphiphilic helical structure and introducing sequence-based short beta-turn motifs.J Biomol Struct Dyn. 2018 Feb;36(2):475-485. doi: 10.1080/07391102.2017.1286265. Epub 2017 Feb 8. J Biomol Struct Dyn. 2018. PMID: 28278764
-
Experimental and computational studies of determinants of membrane-protein folding.Curr Opin Chem Biol. 2002 Dec;6(6):878-84. doi: 10.1016/s1367-5931(02)00393-9. Curr Opin Chem Biol. 2002. PMID: 12470745 Review.
-
[A turning point in the knowledge of the structure-function-activity relations of elastin].J Soc Biol. 2001;195(2):181-93. J Soc Biol. 2001. PMID: 11727705 Review. French.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

