GFI1-driven transcriptional and epigenetic programs maintain CD8+ T cell stemness and persistence
- PMID: 40374731
- PMCID: PMC12133582
- DOI: 10.1038/s41590-025-02151-5
GFI1-driven transcriptional and epigenetic programs maintain CD8+ T cell stemness and persistence
Abstract
Long-lived memory CD8+ T cells are essential for the control of persistent viral infections. The mechanisms that preserve memory cells are poorly understood. Fate mapping of the transcriptional repressor GFI1 identified that GFI1 was differentially regulated in virus-specific CD8+ T cells and was selectively expressed in stem cell memory and central memory cells. Deletion of GFI1 led to reduced proliferation and progressive loss of memory T cells, which in turn resulted in failure to maintain antigen-specific CD8+ T cell populations following infection with chronic lymphocytic choriomeningitis virus or murine cytomegalovirus. Ablation of GFI1 resulted in downregulation of the transcription factors EOMES and BCL-2 in memory CD8+ T cells. Ectopic expression of EOMES rescued the expression of BCL-2, but the persistence of memory CD8+ T cells was only partially rescued. These findings highlight the critical role of GFI1 in the long-term maintenance of memory CD8+ T cells in persistent infections by sustaining their proliferative potential.
© 2025. Crown.
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures










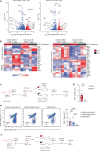
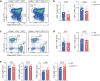
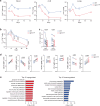



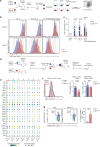
References
-
- Hammarlund, E. et al. Duration of antiviral immunity after smallpox vaccination. Nat. Med.9, 1131–1137 (2003). - PubMed
-
- Utzschneider, D. T. et al. T cell factor 1-expressing memory-like CD8+ T cells sustain the immune response to chronic viral infections. Immunity45, 415–427 (2016). - PubMed
-
- Zehn, D., Thimme, R., Lugli, E., de Almeida, G. P. & Oxenius, A. ‘Stem-like’ precursors are the fount to sustain persistent CD8+ T cell responses. Nat. Immunol.23, 836–847 (2022). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

