The protective effects of propolis against lipopolysaccharide-induced acute liver injury by modulating serum metabolites and gut flora
- PMID: 40374745
- PMCID: PMC12081765
- DOI: 10.1038/s41598-025-01343-1
The protective effects of propolis against lipopolysaccharide-induced acute liver injury by modulating serum metabolites and gut flora
Abstract
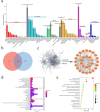
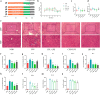
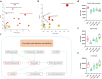
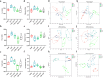
Propolis has significant hepatoprotective effects, but the active components, targets, and mechanisms have not been fully elucidated. Here, we integrated network pharmacology, serum metabolomics, and 16 S rRNA sequencing to disclose the hepatoprotective effects of Chinese propolis (CP) by lipopolysaccharide (LPS)-induced acute liver injury (ALI) in mice. The core active ingredients of CP against ALI, including quercetin, luteolin, and kaempferol, can bind stably to pro-inflammatory factors such as TNF-α, IL-6, IL-1β, and IFN-γ. CP and its active ingredient quercetin obviously alleviated LPS-induced ALI in mice and downregulated the levels of pro-inflammatory genes (Tnf-α, Il-1β, Il-6, Mcp-1, Ifn-γ, and Cox-2) while increasing the protein expression levels of the antioxidant factors Nrf2 and HO-1. Untargeted serum metabolomics analysis indicated that CP and quercetin ameliorated LPS-induced metabolic disorders mainly by modulating the ascorbate and aldarate metabolisms. 16 S rRNA sequencing demonstrated that CP and quercetin modulated the gut microbiota, augmenting the relative abundance of anti-inflammatory bacteria like Lactobacillus and Dubosiella and diminishing the pro-inflammatory bacteria like Alistipes. Spearman correlation analysis revealed that there existed significant correlations among inflammatory factors, gut microbiota, and differential metabolites of serum after propolis pretreatment. Our research indicated that propolis effectively alleviated pathological damage in LPS-induced ALI mice mainly through partially restoring the ecology of gut flora and metabolic disorders to reduce inflammation.
Keywords: Acute liver injury; Chinese propolis; Gut flora; Metabolomics; Network Pharmacology; Quercetin.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Ethics approval: All animal experiments followed the Chinese Guidelines for the Care and Use of Laboratory Animals, which were approved by Liaocheng University’s Special Committee on Scientific Research Ethics (2023103106), and we confirm the study is reported in accordance with ARRIVE guidelines. Conflict of interest: The authors declare that there are no conflicts of interest.
Figures


Similar articles
-
Propolis Alleviates Acute Lung Injury Induced by Heat-Inactivated Methicillin-Resistant Staphylococcus aureus via Regulating Inflammatory Mediators, Gut Microbiota and Serum Metabolites.Nutrients. 2024 May 24;16(11):1598. doi: 10.3390/nu16111598. Nutrients. 2024. PMID: 38892531 Free PMC article.
-
The hepatoprotective effect of the probiotic Clostridium butyricum against carbon tetrachloride-induced acute liver damage in mice.Food Funct. 2017 Nov 15;8(11):4042-4052. doi: 10.1039/c7fo00355b. Food Funct. 2017. PMID: 28933492
-
Integration of microbiomics, metabolomics, and transcriptomics reveals the therapeutic mechanism underlying Fuzheng-Qushi decoction for the treatment of lipopolysaccharide-induced lung injury in mice.J Ethnopharmacol. 2024 Nov 15;334:118584. doi: 10.1016/j.jep.2024.118584. Epub 2024 Jul 15. J Ethnopharmacol. 2024. PMID: 39019418
-
Pharmacodynamic material basis of licorice and mechanisms of modulating bile acid metabolism and gut microbiota in cisplatin-induced liver injury based on LC-MS and network pharmacology analysis.J Ethnopharmacol. 2025 Jan 31;340:119293. doi: 10.1016/j.jep.2024.119293. Epub 2024 Dec 28. J Ethnopharmacol. 2025. PMID: 39736346
-
Therapeutic mechanism of Cornus Officinalis Fruit Coreon on ALI by AKT/Nrf2 pathway and gut microbiota.Phytomedicine. 2024 Jul 25;130:155736. doi: 10.1016/j.phymed.2024.155736. Epub 2024 May 14. Phytomedicine. 2024. PMID: 38788396
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

