Biliary microbiome profiling via 16 S rRNA amplicon sequencing in patients with cholangiocarcinoma, pancreatic carcinoma and choledocholithiasis
- PMID: 40374795
- PMCID: PMC12081727
- DOI: 10.1038/s41598-025-00976-6
Biliary microbiome profiling via 16 S rRNA amplicon sequencing in patients with cholangiocarcinoma, pancreatic carcinoma and choledocholithiasis
Abstract
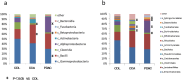
Recent studies have revealed that oral, gut, and intratumoral microbial dysbiosis significantly affects tumor progression, therapy resistance, and prognosis in cholangiocarcinoma (CCA) and pancreatic ductal adenocarcinoma (PDAC) patients. However, the biliary microbiome, which directly interacts with malignant tissues, remains poorly understood. In this study, we analyzed the bile microbiota from 17 CCA, 15 PDAC, and 40 choledocholithiasis (CDL) patients using bacterial 16 S rRNA and fungal ITS sequencing. Principal coordinate analysis revealed significant differences in microbial communities between the cancer and CDL groups. The microbial community structure in each group demonstrated a specific pattern. Linear discriminant analysis revealed Streptococcus, Sphingomonas, and Bacillus enrichment in CCA patients, Neisseria, Sphingomonas, and Caulobacter in PDAC patients were more prevalent compared with CDL patients. Caulobacter was more prevalent, wheares Campylobacter was less in PDAC patients than in CCA patients. Fungal DNA was detected in ~ 50% of the samples, with CCA and PDAC patients. KEGG pathway analysis revealed altered metabolic pathways, including peptidoglycan, sphingolipid, and fatty acid metabolism and bile acid metabolism, in CCA and PDAC patients. These findings highlight the potential role of the biliary microbiome in CCA and PDAC pathogenesis, offering new insights into disease mechanisms and biomarkers.
Keywords: 16S rRNA gene amplicon sequencing; Biliary Microbiome; Cholangiocarcinoma; Choledocholithiasis; Pancreatic carcinoma.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures







Similar articles
-
Profiling of Bile Microbiome Identifies District Microbial Population between Choledocholithiasis and Cholangiocarcinoma Patients.Asian Pac J Cancer Prev. 2021 Jan 1;22(1):233-240. doi: 10.31557/APJCP.2021.22.1.233. Asian Pac J Cancer Prev. 2021. PMID: 33507704 Free PMC article.
-
Dysbiosis of Bile Microbiota in Cholangiocarcinoma Patients: A Comparison with Benign Biliary Diseases.Int J Mol Sci. 2025 Feb 13;26(4):1577. doi: 10.3390/ijms26041577. Int J Mol Sci. 2025. PMID: 40004041 Free PMC article.
-
Distinct composition and metabolic potential of biliary microbiota in patients with malignant bile duct obstruction.Eur J Gastroenterol Hepatol. 2025 May 1;37(5):585-593. doi: 10.1097/MEG.0000000000002948. Epub 2025 Mar 26. Eur J Gastroenterol Hepatol. 2025. PMID: 40207470 Free PMC article.
-
Association of Chronic Opisthorchis Infestation and Microbiota Alteration on Tumorigenesis in Cholangiocarcinoma.Clin Transl Gastroenterol. 2020 Dec 22;12(1):e00292. doi: 10.14309/ctg.0000000000000292. Clin Transl Gastroenterol. 2020. PMID: 33464733 Free PMC article. Review.
-
Role of Gut Microbial Metabolites in the Pathogenesis of Primary Liver Cancers.Nutrients. 2024 Jul 22;16(14):2372. doi: 10.3390/nu16142372. Nutrients. 2024. PMID: 39064815 Free PMC article. Review.
References
-
- Thomas, R. M. & Jobin, C. Microbiota in pancreatic health and disease: the next frontier in Microbiome research. Nat. Rev. Gastroenterol. Hepatol. Jan. 17 (1), 53–64 (2020). - PubMed
-
- Nagata, N. et al. Metagenomic identification of microbial signatures predicting pancreatic Cancer from a multinational study. GastroenterologyJul;163 (1), 222–238 (2022). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

