Hybridization of SMILES and chemical-environment-aware tokens to improve performance of molecular structure generation
- PMID: 40374848
- PMCID: PMC12081657
- DOI: 10.1038/s41598-025-01890-7
Hybridization of SMILES and chemical-environment-aware tokens to improve performance of molecular structure generation
Abstract
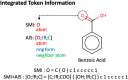
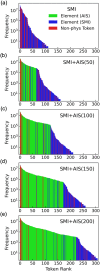
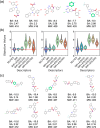
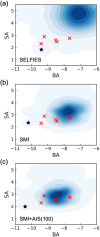
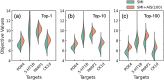
The Simplified Molecular Input Line Entry System (SMILES) is one of the most widely adopted molecular representations. However, SMILES notation suffers from limited token diversity and a lack of chemical information within individual tokens. To address these limitations while maintaining its simplicity, we propose a molecular representation method through the hybridization of standard SMILES tokens with Atom-In-SMILES (AIS) tokens, which incorporate local chemical environment information into a single token. This hybrid representation, termed SMI + AIS, combines SMILES and AIS tokens, allowing AIS tokens to differentiate chemical elements based on their chemical context without introducing additional tokens for less frequent elements. Using the SMI + AIS representation, we evaluated its performance by comparing the predefined metric of generated structures in chemical structure generation based on latent space optimization. Compared to standard SMILES, SMI + AIS achieved a 7% improvement in binding affinity and a 6% increase in synthesizability, highlighting its utility in the enhancement of machine learning-based molecular design. Our results demonstrate that the SMI + AIS representation provides a more effective and informative approach to encapsulate chemical context and presents potential for performance enhancement in other machine learning tasks in chemistry.
Keywords: Drug discovery; Generative modeling; Latent space optimization; Molecular representation; SMILES; Small-molecule drug.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures

References
-
- Fialkowski, M. et al. Architecture and evolution of organic chemistry. Angew. Chem. Int. Ed.44, 7263–7269. 10.1002/anie.200502272 (2005). - PubMed
-
- Lipkus, A. H. et al. Recent changes in the scaffold diversity of organic chemistry as seen in the CAS registry. J. Org. Chem.84, 13948–13956. 10.1021/acs.joc.9b02111 (2019). - PubMed
-
- Coley, C. W. et al. Convolutional embedding of attributed molecular graphs for physical property prediction. J. Chem. Inf. Model57, 1757–1772. 10.1021/acs.jcim.6b00601 (2017). - PubMed
-
- Guo, Z., Guo, K., Nan, B. et al. Graph-Based Molecular Representation Learning (2022). 10.48550/ARXIV.2207.04869
Grants and funding
LinkOut - more resources
Full Text Sources

