A combined in silico and MD simulation approach to discover novel LpxC inhibitors targeting multiple drug resistant Pseudomonas aeruginosa
- PMID: 40374903
- PMCID: PMC12081860
- DOI: 10.1038/s41598-025-99215-1
A combined in silico and MD simulation approach to discover novel LpxC inhibitors targeting multiple drug resistant Pseudomonas aeruginosa
Abstract
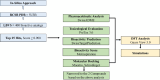
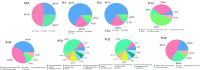
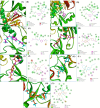
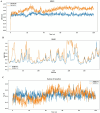
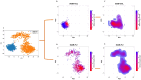
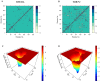
Pseudomonas aeruginosa (P. aeruginosa), a member of the ESKAPE family, is the major cause of infections leading to increased morbidity and mortality due to multidrug resistance (MDR). One of the main proteins involved in the Raetz pathway is LpxC, which plays a significant role in anti-microbial resistance (AMR). Our study aimed to identify a novel compound to combat MDR due to the LpxC protein. It involved in silico methods comprising molecular docking, simulations, ADMET profiling, and DFT calculations. First, an ADMET and bioactivity evaluation of the 25 top-hit compounds retrieved from ligand-based virtual screening was performed, followed by molecular docking. The results revealed compound P-2 as the lead compound, which was further subjected to DFT analysis and molecular dynamics (MD) simulations. With these analyses, our in silico study identified P-2, 3-[(dimethylamino)methyl]-N-[(2 S)-1-(hydroxyamino)-1-oxobutan-2-yl]benzamide as a potential lead compound that may behave as a very potent inhibitor of LpxC for the development of targeted therapies against MDR P. aeruginosa.
Keywords: ADMET; Bioactivity; DFT; In silico; LpxC; MD simulation; MDR; Molecular docking; Toxicity.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures
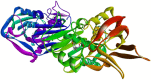


References
-
- Amera, G. M. et al. Computer aided ligand based screening for identification of promising molecules against enzymes involved in peptidoglycan biosynthetic pathway from Acinetobacter baumannii. Microb. Pathog.147, 104205 (2020). - PubMed
-
- Tacconelli, E. et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis.18, 318–327 (2018). - PubMed
-
- Shevade, A. & Naik, S. Mitigation of antimicrobial resistance (AMR) in G20. Indian Public. Policy Rev.4, 1–29 (2023).
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

