Deciphering the role of acetylation-related gene NAT10 in colon cancer progression and immune evasion: implications for overcoming drug resistance
- PMID: 40374962
- PMCID: PMC12081795
- DOI: 10.1007/s12672-025-02617-w
Deciphering the role of acetylation-related gene NAT10 in colon cancer progression and immune evasion: implications for overcoming drug resistance
Abstract
Background: Colon cancer (CC) is one of the most common and lethal cancers worldwide, with rising incidence rates in both developed and developing countries. Although advances in treatments such as surgery, chemotherapy, and targeted therapies have been made, prognosis for advanced colon cancer, particularly with metastasis, remains poor. Recent studies highlight the significant role of post-transcriptional modifications like acetylation in cancer biology, affecting processes like gene transcription, metabolism, and tumor progression.
Methods: This study applied multi-omics analyses, including single-cell RNA sequencing (scRNA-seq), spatial transcriptomics, and Mendelian randomization. Data were obtained from public datasets like GSE132465, UCSC Xena, and GeneCards. We focused on acetylation-related genes, specifically NAT10 and GNE, using scoring methods, cell-cell interaction models, and survival analyses to investigate their role in colon cancer development, metastasis, and immune evasion.
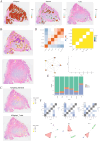
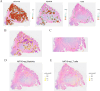
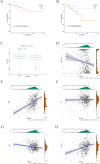
Results: This study identifies that NAT10 is highly expressed in epithelial cells of colorectal cancer (CC) and is closely associated with tumor progression and metastasis. Single-cell RNA sequencing analysis revealed that NAT10-positive epithelial cells exhibited strong interactions with myeloid cells and T cells, with significant differences in cell-cell communication (p < 0.05). Based-on-summary-data Mendelian randomization (SMR) analysis further supports a causal relationship between NAT10 and colorectal cancer. In the MR analysis, a significant positive correlation was observed between NAT10 and colorectal cancer risk using summary data from genome-wide association studies (GWAS) and expression quantitative trait loci (eQTL) studies (β_SMR = 0.004, p_SMR = 0.041, p_HEIDI = 0.737). These findings suggest that NAT10 may serve as a pathogenic factor in colorectal cancer development, providing additional genetic evidence that links this acetylation-related gene to colorectal cancer. Survival analysis further demonstrated that NAT10-positive epithelial cells are associated with poorer prognosis. In the TCGA dataset, patients with NAT10-positive epithelial cells exhibited a significantly shorter disease-free survival (DFS) (p = 0.012). Unlike GNE-positive cells, NAT10-positive epithelial cells exhibited immune escape characteristics, and TIDE analysis indicated that NAT10-positive epithelial cells were associated with a lower response to immune checkpoint blockade therapy (p = 1.3e-5), suggesting that they may impair the efficacy of immunotherapy by promoting immune evasion. In contrast, GNE was also significantly expressed in epithelial cells of colorectal cancer, but its role differs from that of NAT10. GNE-positive epithelial cells demonstrated strong communication with immune cells, particularly in interactions between myeloid cells and T cells through receptor-ligand pairs. Despite the important role of GNE-positive epithelial cells in the tumor microenvironment, their association with immune escape is weaker compared to NAT10. Survival analysis revealed that GNE-positive epithelial cells were associated with a better prognosis (p = 0.015). In the TCGA dataset, patients with GNE-positive epithelial cells displayed longer disease-free survival (DFS), contrary to the results from the SMR analysis.
Conclusions: Leveraging SMR and multi-omics analysis, this study highlights the significant role of acetylation-related genes, particularly NAT10, in colon cancer. The findings suggest that acetylation modifications in epithelial cells contribute to immune evasion and cancer progression. NAT10 could serve as a promising biomarker and therapeutic target for early diagnosis and targeted therapy, offering new avenues for improving colon cancer treatment and patient outcomes.
Keywords: Acetylation; Colon cancer; GNE; Immune evasion; Multi-omics analysis; NAT10; Prognosis; Single-cell RNA sequencing; Spatial transcriptomics; Tumor microenvironment.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Conflict of interests: The authors declare no competing interests.
Figures





Similar articles
-
The association between immune cells and breast cancer: insights from Mendelian randomization and meta-analysis.Int J Surg. 2025 Jan 1;111(1):230-241. doi: 10.1097/JS9.0000000000001840. Int J Surg. 2025. PMID: 38935111 Free PMC article.
-
NAT10 mediated mRNA acetylation modification patterns associated with colon cancer progression and microsatellite status.Epigenetics. 2023 Dec;18(1):2188667. doi: 10.1080/15592294.2023.2188667. Epigenetics. 2023. PMID: 36908042 Free PMC article.
-
N-acetyltransferase 10 promotes colon cancer progression by inhibiting ferroptosis through N4-acetylation and stabilization of ferroptosis suppressor protein 1 (FSP1) mRNA.Cancer Commun (Lond). 2022 Dec;42(12):1347-1366. doi: 10.1002/cac2.12363. Epub 2022 Oct 8. Cancer Commun (Lond). 2022. PMID: 36209353 Free PMC article.
-
The role and mechanism of NAT10-mediated ac4C modification in tumor development and progression.MedComm (2020). 2024 Dec 4;5(12):e70026. doi: 10.1002/mco2.70026. eCollection 2024 Dec. MedComm (2020). 2024. PMID: 39640362 Free PMC article. Review.
-
Emerging Role of NAT10 as ac4C Writer in Inflammatory Diseases: Mechanisms and Therapeutic Applications.Curr Drug Targets. 2025;26(4):282-294. doi: 10.2174/0113894501346709241202110834. Curr Drug Targets. 2025. PMID: 39633518 Review.
Cited by
-
The Key Role of COA6 in Pancreatic Ductal Adenocarcinoma: Metabolic Reprogramming and Regulation of the Immune Microenvironment.J Cell Mol Med. 2025 Jul;29(13):e70685. doi: 10.1111/jcmm.70685. J Cell Mol Med. 2025. PMID: 40596639 Free PMC article.
-
Joint exposure to PM2.5, warm-season heat, and sedentary behavior accelerates incident lung cancer in ageing Chinese adults: evidence from CHARLS.Front Public Health. 2025 Jul 8;13:1622767. doi: 10.3389/fpubh.2025.1622767. eCollection 2025. Front Public Health. 2025. PMID: 40697836 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
