GJB2 as a novel prognostic biomarker associated with immune infiltration and cuproptosis in ovarian cancer
- PMID: 40375037
- PMCID: PMC12167356
- DOI: 10.1007/s10495-025-02119-8
GJB2 as a novel prognostic biomarker associated with immune infiltration and cuproptosis in ovarian cancer
Abstract
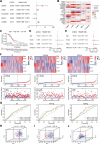
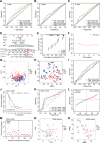
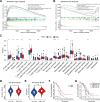
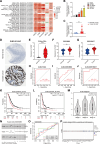
Cuproptosis, a recently identified copper-dependent cell death mechanism, remains poorly unexplored in ovarian cancer (OC). This study systematically evaluates clinically significant cuproptosis-related genes (CRGs) as potential prognostic biomarkers in OC. Cox regression analysis and LASSO algorithms were used to develop a prognostic risk model incorporating 5 CRGs (CD8B2, GJB2, GRIP2, MELK, and PLA2G2D) within the TCGA cohort. This model stratified OC patients into high-risk and low-risk groups, with the high-risk group exhibiting significantly shorter overall survival compared to the low-risk group. The model's predictive accuracy for prognosis in OC patients was validated in the TCGA training cohort, TCGA testing cohort, and ICGC external validation cohorts. Among these 5 signature genes, the number of cuproptosis genes associated with GJB2 is the largest, so we selected GJB2 for further validation. qPCR revealed that GJB2 was highly expressed in OC cells and tumor tissues. The high expression of GJB2 was closely associated with poor prognosis in OC patients. Functionally, GJB2 silencing suppressed OC cell proliferation and migration while its overexpression promoted malignant progression and EMT. Furthermore, GJB2 regulated copper homeostasis and reduced cuproptosis sensitivity, while also facilitating immune escape by inhibiting CD8+ T cell infiltration and cytokine secretion, revealing its multiple roles in OC progression. In conclusion, we established a novel prognostic model incorporating 5 CRGs that effectively predicts clinical outcomes and characterizes the immune microenvironment in OC. Our findings particularly highlight GJB2 as a key regulator of cuproptosis with significant potential as both a prognostic biomarker and therapeutic target for OC management.
Keywords: GJB2; Cuproptosis; Immune infiltration; Ovarian cancer; Prognostic signature; Proliferation and migration.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Ethics approval: The study was reviewed and approved by the Ethics Committee of Xiangya Hospital Central South University, approval number 202204081. All clinical samples were collected with informed consent from patients. We declare that all methods are reported in accordance with the declaration of Helsinki.
Figures





Similar articles
-
Interplay between tumor mutation burden and the tumor microenvironment predicts the prognosis of pan-cancer anti-PD-1/PD-L1 therapy.Front Immunol. 2025 Jul 24;16:1557461. doi: 10.3389/fimmu.2025.1557461. eCollection 2025. Front Immunol. 2025. PMID: 40777041 Free PMC article.
-
Prognostic and Immunological Significance of the Molecular Subtypes and Risk Signatures Based on Cuproptosis in Hepatocellular Carcinoma.Mediators Inflamm. 2023 Apr 20;2023:3951940. doi: 10.1155/2023/3951940. eCollection 2023. Mediators Inflamm. 2023. PMID: 37124062 Free PMC article.
-
Construction and validation of a lipid metabolism-related genes prognostic signature for skin cutaneous melanoma.Biochem Biophys Res Commun. 2025 Aug 15;775:152115. doi: 10.1016/j.bbrc.2025.152115. Epub 2025 May 29. Biochem Biophys Res Commun. 2025. PMID: 40460484
-
Impact of residual disease as a prognostic factor for survival in women with advanced epithelial ovarian cancer after primary surgery.Cochrane Database Syst Rev. 2022 Sep 26;9(9):CD015048. doi: 10.1002/14651858.CD015048.pub2. Cochrane Database Syst Rev. 2022. PMID: 36161421 Free PMC article.
-
Systemic treatments for metastatic cutaneous melanoma.Cochrane Database Syst Rev. 2018 Feb 6;2(2):CD011123. doi: 10.1002/14651858.CD011123.pub2. Cochrane Database Syst Rev. 2018. PMID: 29405038 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

