Genome-wide identification, tissue expression pattern, and salt stress response analysis of the NAC gene family in Thinopyrum elongatum
- PMID: 40375071
- PMCID: PMC12080115
- DOI: 10.1186/s12870-025-06696-3
Genome-wide identification, tissue expression pattern, and salt stress response analysis of the NAC gene family in Thinopyrum elongatum
Abstract
Background: The NAC (NAM, ATAF1/2, and CUC2) gene family plays a critical role in regulating plant growth, developmental processes, and stress response mechanisms. While NAC genes have been systematically characterized in multiple plant species, this study focused on genome-wide identification of NAC family members in Thinopyrum elongatum (designated as TeNACs) through integrated bioinformatics approaches. Comprehensive analyses were conducted to determine the physicochemical characteristics, conserved motifs, gene structure, phylogenetic relationships, chromosomal collinearity, and expression profiles of the identified TeNACs. This multi-dimensional characterization provides fundamental insights into the structural and functional diversity of NAC transcription factors in Th. elongatum.
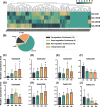
Results: A total of 116 NAC transcription factors were systematically identified in Th. elongatum, distributed across all seven chromosomes. Comprehensive physicochemical characterization revealed substantial variation among TeNAC proteins: amino acid length (162-718 aa), molecular weight (18.33-78.25 kDa), isoelectric point (6.5-7.5), instability index (30.34-63.32), and aliphatic index (50.92-80.68). Hydropathicity analysis confirmed the hydrophilic nature of all TeNACs, with grand average values consistently below zero. Conserved motif profiling demonstrated a highly conserved architecture in TeNACs, featuring ordered arrangements of motifs 3, 4, 1, 5, 6, 2, and 7. Phylogenetic reconstruction classified TeNACs into 14 distinct clades through comparative analysis with Arabidopsis thaliana NAC genes, notably lacking ANAC001 and OsNAC8 homologs. Comparative genomic analysis identified significant syntenic conservation between TeNACs and wheat NAC genes. Protein interaction network prediction indicated intricate functional associations among TeNAC proteins. Computational predictions coupled with experimental validation of TeNAC021 confirmed exclusive nuclear localization for all family members. Differential expression analysis across a salt gradient (0-300 mM) identified 14 TeNACs with progressive up-regulation and 5 showing consistent down-regulation. RT-qPCR confirmed salt-responsive expression patterns for eight TeNACs demonstrating marked transcriptional changes.
Conclusions: This systematic investigation establishes a robust theoretical framework for subsequent structural and functional characterization of TeNACs in Th. elongatum.
Keywords: TeNACs; Th. elongatum; Expression pattern; Salt stress.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures



Similar articles
-
Genome-wide analysis of the Tritipyrum NAC gene family and the response of TtNAC477 in salt tolerance.BMC Plant Biol. 2024 Jan 9;24(1):40. doi: 10.1186/s12870-023-04629-6. BMC Plant Biol. 2024. PMID: 38195389 Free PMC article.
-
Genome-wide investigation and analysis of NAC transcription factor family in Populus tomentosa and expression analysis under salt stress.Plant Biol (Stuttg). 2024 Aug;26(5):764-776. doi: 10.1111/plb.13657. Epub 2024 Jun 10. Plant Biol (Stuttg). 2024. PMID: 38859551
-
Genome-wide analysis of the Tritipyrum bHLH gene family and the response of TtbHLH310 in salt-tolerance.BMC Genomics. 2025 May 30;26(1):549. doi: 10.1186/s12864-025-11657-z. BMC Genomics. 2025. PMID: 40448022 Free PMC article.
-
Genome-Wide Identification and Analysis of the NAC Transcription Factor Gene Family in Garden Asparagus (Asparagus officinalis).Genes (Basel). 2022 May 30;13(6):976. doi: 10.3390/genes13060976. Genes (Basel). 2022. PMID: 35741738 Free PMC article.
-
Genome-wide identification, characteristics and expression of the prolamin genes in Thinopyrum elongatum.BMC Genomics. 2021 Dec 2;22(1):864. doi: 10.1186/s12864-021-08088-x. BMC Genomics. 2021. PMID: 34852761 Free PMC article. Review.
References
-
- Souer E, van Houwelingen A, Kloos D, Mol J, Koes R. The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell. 1996;85(2):159–70. - PubMed
-
- Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P, et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003;10(6):239–47. - PubMed
-
- Zhu T, Nevo E, Sun D, Peng J. Phylogenetic analyses unravel the evolutionary history of NAC proteins in plants. Evolution. 2012;66(6):1833–48. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

