Comparative transcriptome analysis reveals key genes associated with meiotic stability and high seed setting rate in tetraploid rice
- PMID: 40375090
- PMCID: PMC12080012
- DOI: 10.1186/s12870-025-06672-x
Comparative transcriptome analysis reveals key genes associated with meiotic stability and high seed setting rate in tetraploid rice
Abstract
Background: Polyploid rice has a high yield potential and excellent nutritional quality. The development of polyploid rice remained critically limited for several decades due to low seed setting rate until the successful breeding of polyploid meiosis stability (PMeS) lines. To determine the mechanism responsible for meiotic stability and high seed setting rate of PMeS line, agronomic traits, pollen fertility and viability, and meiotic behaviors of PMeS and non-PMeS lines were investigated. Further, comparative transcriptome analysis was performed to identify genes associated with meiotic stability and high seed setting rate in PMeS line.
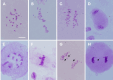
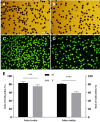
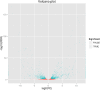
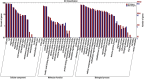
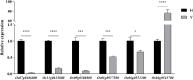
Results: The seed setting rate, fertile and viable pollen ratios of PMeS line were significantly higher than those of non-PMeS line. The PMeS line exhibited stable meiosis, and chromosomes mainly paired as bivalents, rarely as univalents and multivalents in prophase I. Few lagging chromosomes were observed in anaphase I. By contrast, the homologous chromosomes pairing was disorganized in the non-PMeS line, with low frequencies of bivalents and high frequencies of univalents and multivalents in prophase I, while more cells with increased lagging chromosomes were detected in anaphase I. Many differentially expressed genes (DEGs) between PMeS and non-PMeS lines were identified through comparative transcriptome analysis. Some meiosis-related genes were specifically investigated from all DEGs. Further, several meiotic genes were identified as candidate genes.
Conclusions: The study not only demonstrates the morphological, cytological, and molecular differences between the PMeS and non-PMeS lines, but also provides several key genes associated with meiotic stability and high seed setting rate in tetraploid rice.
Keywords: High seed setting rate; Meiotic genes; Meiotic stability; Tetraploid rice; Transcriptome analysis.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures

References
-
- Comai L. The advantages and disadvantages of being polyploid. Nat Rev Genet. 2005;6:836–46. - PubMed
-
- Soltis DE, Albert VA, Leebens-Mack J, Bell CD, Paterson AH, Zheng C, et al. Polyploidy and angiosperm diversification. Am J Bot. 2009;96:336–48. - PubMed
-
- Fang Z, Morrell PL. Domestication: polyploidy boosts domestication. Nat Plants. 2016;2:16116. - PubMed
-
- Yu H, Lin T, Meng X, Du H, Zhang J, Liu G, et al. A route to de Novo domestication of wild allotetraploid rice. Cell. 2021;184:1156–70. - PubMed
-
- Otto SP. The evolutionary consequences of polyploidy. Cell. 2007;131:452–62. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

