TREAT: systematic and inclusive selection process of genes for genomic newborn screening as part of the Screen4Care project
- PMID: 40375093
- PMCID: PMC12082943
- DOI: 10.1186/s13023-025-03692-6
TREAT: systematic and inclusive selection process of genes for genomic newborn screening as part of the Screen4Care project
Abstract
Background: Genomic newborn screening (gNBS) offers the potential to detect genetic conditions early, enhancing outcomes through timely treatment. It can serve as an additional tool to identify conditions that are not detectable via metabolic screening. The Screen4Care project seeks to develop a systematic approach for selecting treatable rare diseases (RDs) for inclusion in gNBS through the creation of the TREAT-panel.
Methods: A set of six selection criteria containing treatability, clinical validity, age of onset, disease severity, penetrance, and genetic feasibility was applied to a comprehensive list of gene-disease pairs. Genes meeting a defined threshold score were included in the TREAT-panel. This automated scoring process was complemented by expert review from clinicians and patient representatives to ensure clinical relevance and adherence to current medical guidelines.
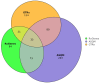
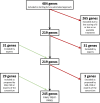
Results: The initial gene list, derived from multiple data sources, included 484 gene-disease pairs. After applying the scoring system and two rounds of expert curation, a final list of 245 genes was selected. These genes predominantly represent disorders in metabolic, neurological, and immunological categories, with treatability and early disease onset as key inclusion factors.
Conclusion: The Screen4Care TREAT-panel provides a curated, scientifically robust gene set for gNBS, focusing on treatable RDs with early onset and clinical actionability. The panel will be tested in a European pilot project involving approximately 20,000 newborns, contributing to the growing body of evidence for the implementation of next-generation sequencing (NGS) in newborn screening programs.
Keywords: Diagnosis; Genetic; Newborn screening; Paediatric; Rare diseases; Treatable diseases.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The disease selection process described in this manuscript did not involve any experiments with human subjects. The planed pilot project for genetic newborn screening with the TREAT-panel gene list has been approved by the Freiburg University Ethics Committee in Germany (24-1084_1-S1), the Italian Ethics Committee of the University of Ferrara (758/2023/Sper/AOUFe) and the French Ethics Committee (24.01886.000348). Consent for publication: Not applicable. Competing interests: EB has received funds for Advisory Board from Roche, Biogen, PTC, Pfizer. JK has received funds for Advisory Boards from Biogen, Novartis, Pfizer, Roche, Santhera and research funding from Biogen, Novartis and Roche. SS is an employee of Sanofi. JB is an employee of Pfizer. ME and YE are employees of Genoox. All the other authors declare that they have no competing interests. Financial and non-financial interests: EB has received funds for Advisory Board from Roche, Biogen, PTC, Pfizer. JK has received funds for Advisory Boards from Biogen, Novartis, Pfizer, Roche, Santhera and research funding from Biogen, Novartis and Roche. SS is an employee of Sanofi. JB is an employee of Pfizer. ME and YE are employees of Genoox. NG was a full-time employee of Pfizer Inc. at the beginning of this work and is now a full-time employee of Servier Affaires Médicales. All the other authors declare that they have no competing interests.
Figures
References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

