Genome-wide enhancer-gene regulatory maps of liver reveal novel regulatory mechanisms underlying NAFLD pathogenesis
- PMID: 40375105
- PMCID: PMC12082939
- DOI: 10.1186/s12864-025-11668-w
Genome-wide enhancer-gene regulatory maps of liver reveal novel regulatory mechanisms underlying NAFLD pathogenesis
Abstract
Introduction: Non-alcoholic fatty liver disease (NAFLD) represents the most widespread liver disease globally, ranging from non-alcoholic fatty liver (NAFL) and steatohepatitis (NASH) to fibrosis/cirrhosis, with potential progression to hepatocellular carcinoma (HCC). Genome-wide association studies (GWASs) have identified several single nucleotide polymorphisms (SNPs) associated with NAFLD. However, numerous GWAS signals associated with NAFLD locate in non-coding regions, posing a challenge for interpreting their functional annotation.
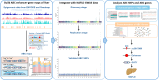
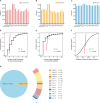
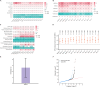
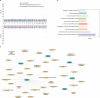
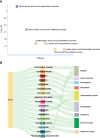
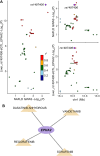
Results: In this study, we utilized the Activity-by-Contact (ABC) model to construct the enhancer-gene maps of liver by integrating epigenomic data from 15 liver tissues and cell lines. We constructed the most comprehensive genome-wide regulatory maps of the liver, identifying 543,486 enhancer-gene connections, including 267,857 enhancers and 16,872 target genes. Enrichment analyses revealed that the ABC SNPs are significantly enriched in active chromatin regions and active chromatin state. By combining the ABC regulatory maps and NAFLD GWAS data, we systematically identified ABC SNPs associated with NAFLD risk. Through the functional annotations, such as pathway enrichment and drug-gene interaction analyses, we identified 6 genes (GGT1, ACTG1, SPP1, EPHA2, PROZ and SHMT1) as candidate NAFLD genes, with SHMT1 previously reported. Among the SNPs connected to the candidate genes, the ABC SNP rs2017869 (odds ratio [OR] for the C allele = 1.10, 95% CI = 1.04-1.16, P = 5.97 × 10- 4) had the highest ABC score. According to the ABC maps, rs2017869 links to GGT1, and several drugs targeting this gene, such as liothyronine, showed potential benefits to patients with NAFLD. Furthermore, we identified that another novel gene, EPHA2, may play a crucial role in NAFLD by regulating the GGT levels.
Conclusions: Our study provides the most comprehensive ABC regulatory maps of the liver to date. This resource offers a valuable reference for identifying regulatory variants and prioritizing susceptibility genes of liver diseases, such as NAFLD.
Keywords: GGT1; Genome-wide association study; Non-alcoholic fatty liver disease; Single nucleotide polymorphism.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Ethical approval was obtained from the original studies. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures
Similar articles
-
Genome-wide association study of non-alcoholic fatty liver and steatohepatitis in a histologically characterised cohort☆.J Hepatol. 2020 Sep;73(3):505-515. doi: 10.1016/j.jhep.2020.04.003. Epub 2020 Apr 13. J Hepatol. 2020. PMID: 32298765
-
Bioinformatics analysis reveals novel core genes associated with nonalcoholic fatty liver disease and nonalcoholic steatohepatitis.Gene. 2020 Jun 5;742:144549. doi: 10.1016/j.gene.2020.144549. Epub 2020 Mar 14. Gene. 2020. PMID: 32184169
-
Integrative analysis of liver-specific non-coding regulatory SNPs associated with the risk of coronary artery disease.Am J Hum Genet. 2021 Mar 4;108(3):411-430. doi: 10.1016/j.ajhg.2021.02.006. Epub 2021 Feb 23. Am J Hum Genet. 2021. PMID: 33626337 Free PMC article.
-
NAFLD: genetics and its clinical implications.Clin Res Hepatol Gastroenterol. 2022 Nov;46(9):102003. doi: 10.1016/j.clinre.2022.102003. Epub 2022 Aug 11. Clin Res Hepatol Gastroenterol. 2022. PMID: 35963605 Review.
-
The genetic backgrounds in nonalcoholic fatty liver disease.Clin J Gastroenterol. 2018 Apr;11(2):97-102. doi: 10.1007/s12328-018-0841-9. Epub 2018 Feb 28. Clin J Gastroenterol. 2018. PMID: 29492830 Review.
References
-
- Younossi Z, Anstee QM, Marietti M, Hardy T, Henry L, Eslam M, George J, Bugianesi E. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol. 2018;15(1):11–20. - PubMed
-
- Riazi K, Azhari H, Charette JH, Underwood FE, King JA, Afshar EE, Swain MG, Congly SE, Kaplan GG, Shaheen AA. The prevalence and incidence of NAFLD worldwide: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol. 2022;7(9):851–61. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

