New insights into Eragrostis curvula's sexual and apomictic reproductive development
- PMID: 40376162
- PMCID: PMC12078246
- DOI: 10.3389/fpls.2025.1530855
New insights into Eragrostis curvula's sexual and apomictic reproductive development
Abstract
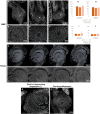
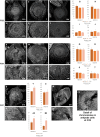
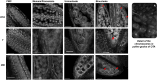
Apomixis, defined as asexual propagation by seeds, is considered of great importance for agriculture as it allows the fixation of desired traits and its propagation through generations. Eragrostis curvula (Schrad.) Ness, is a perennial grass that comprises a polymorphic complex including sexual and diplosporous apomictic cytotypes, where all apomicts are polyploids. In this study we present the first detailed description of female and male gametophyte development in E. curvula through confocal laser microscopy, contrasting three genotypes: the fully apomictic Tanganyika, the facultative apomictic Don Walter, and the sexual OTA-S. Moreover, we have studied the localized expression of a gene known as SQUAMOSA PROMOTER BINDING PROTEIN-LIKE7 (SPL7), that was found to be differentially expressed in contrasting genotypes of E. curvula. This gene had been previously linked with flower development and abiotic stresses in several species, thus, in situ hybridizations were carried out in the model plant Arabidopsis thaliana, as well as in sexual and apomictic E. curvula genotypes. Our microscopy analysis has led to the identification of specific morphological characteristics for each genotype, mainly depicting a larger ovule in the sexual genotype's reproductive development after the meiosis stage. These results reveal potentially important features, which could be used for a simple identification of genotypes. Moreover, differential expression of the gene SPL7 was detected, specifically determining an overexpression of the gene in the sexual genotype. These results demonstrated that it could be an interesting candidate to understand the mechanisms behind apomictic development.
Keywords: Eragrostis curvula; apomixis; confocal laser microscopy; ovule development; pollen development; sexual reproduction.
Copyright © 2025 Pasten, Carballo, Díaz, Mizzotti, Cucinotta, Colombo, Echenique and Mendes.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
A combined transcriptome - miRNAome approach revealed that a kinesin gene is differentially targeted by a novel miRNA in an apomictic genotype of Eragrostis curvula.Front Plant Sci. 2022 Sep 30;13:1012682. doi: 10.3389/fpls.2022.1012682. eCollection 2022. Front Plant Sci. 2022. PMID: 36247597 Free PMC article.
-
A sexual/apomictic consensus linkage map of Eragrostis curvula at tetraploid level.BMC Plant Biol. 2025 May 17;25(1):658. doi: 10.1186/s12870-025-06676-7. BMC Plant Biol. 2025. PMID: 40382602 Free PMC article.
-
Characterization and discovery of miRNA and miRNA targets from apomictic and sexual genotypes of Eragrostis curvula.BMC Genomics. 2019 Nov 12;20(1):839. doi: 10.1186/s12864-019-6169-0. BMC Genomics. 2019. PMID: 31718556 Free PMC article.
-
Eragrostis curvula, a Model Species for Diplosporous Apomixis.Plants (Basel). 2021 Aug 31;10(9):1818. doi: 10.3390/plants10091818. Plants (Basel). 2021. PMID: 34579351 Free PMC article. Review.
-
Apomixis in plant reproduction: a novel perspective on an old dilemma.Plant Reprod. 2013 Sep;26(3):159-79. doi: 10.1007/s00497-013-0222-y. Epub 2013 Jul 14. Plant Reprod. 2013. PMID: 23852378 Free PMC article. Review.
References
-
- Aron Y., Gazit S., Czosnek H., Degani C. (1998). Polyembryony in mango (Mangifera indica L.) is controlled by a single dominant gene. HortScience 33, 1241–1242. doi: 10.21273/HORTSCI.33.7.1241 - DOI
-
- Asker S., Jerling L. (1992). Apomixis in plants (Boca Raton, Florida: CRC Press, Inc.), ISBN: ISBN 0-8493-4545-6.

