Development of A Novel EvaGreen-Dye Based Recombinase Aided Amplification Assay Using Self-Avoiding Molecular Recognition System Primers
- PMID: 40376447
- PMCID: PMC12075452
- DOI: 10.46234/ccdcw2025.079
Development of A Novel EvaGreen-Dye Based Recombinase Aided Amplification Assay Using Self-Avoiding Molecular Recognition System Primers
Abstract
Introduction: Fluorescent probe-based recombinase aided amplification (RAA) offers the advantages of rapidity and simplicity but is limited by the requirement for complex and lengthy probe design, restricting its widespread application.
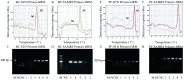
Methods: A novel EvaGreen dye-based RAA (EvaGreen-RAA) assay utilizing self-avoiding molecular recognition system (SAMRS) primers was developed for the detection of Pseudomonas fluorescens (PF) and Bacillus cereus (BC) in milk. Conventional RAA was used as a reference method. Sensitivity was evaluated using nucleic acids from recombinant plasmids and simulated milk specimens. Additionally, a dual EvaGreen-RAA assay was investigated for simultaneous detection of mixed BC and PF in simulated milk specimens.
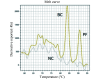
Results: The EvaGreen-RAA demonstrated superior sensitivity compared to conventional RAA, with detection limits of 1 copy/µL versus 10 copies/µL for both BC and PF plasmids, respectively. In simulated milk specimens, EvaGreen-RAA detected BC and PF at concentrations of 100 CFU/mL and 200 CFU/mL, respectively, compared to 400 CFU/mL and 600 CFU/mL for conventional RAA. The dual EvaGreen-RAA assay successfully detected mixed BC and PF in simulated milk specimens at concentrations of 200 CFU/mL for each pathogen.
Conclusion: The EvaGreen-RAA assay demonstrated significant advantages in terms of simplicity and enhanced sensitivity compared to fluorescent probe-based RAA, offering a novel approach for developing multiplex pathogen detection systems using melting curve analysis.
Keywords: Bacillus cereus; EvaGreen dye; Melt Curve; Pseudomonas fluorescens; Raw milk; Recombinase aided amplification; SAMRS.
Copyright © 2025 by Chinese Center for Disease Control and Prevention.
Conflict of interest statement
No conflicts of interest.
Figures

Similar articles
-
[Establishment and preliminary application of a recombinase-aided isothermal amplification assay-based multiplex nucleic acid assay for detection of three Echinococcus species].Zhongguo Xue Xi Chong Bing Fang Zhi Za Zhi. 2021 Aug 19;33(4):339-345. doi: 10.16250/j.32.1374.2021094. Zhongguo Xue Xi Chong Bing Fang Zhi Za Zhi. 2021. PMID: 34505439 Chinese.
-
[Establishment of a nucleic acid assay for detection of Echinococcus granulosus based on recombinase-aided isothermal amplification assay].Zhongguo Xue Xi Chong Bing Fang Zhi Za Zhi. 2020 Jul 2;32(4):340-344. doi: 10.16250/j.32.1374.2020071. Zhongguo Xue Xi Chong Bing Fang Zhi Za Zhi. 2020. PMID: 32935505 Chinese.
-
Development and evaluation of a sensitive recombinase aided amplification assay for rapid detection of Vibrio parahaemolyticus.J Microbiol Methods. 2022 Feb;193:106404. doi: 10.1016/j.mimet.2021.106404. Epub 2022 Jan 3. J Microbiol Methods. 2022. PMID: 34990645
-
Advances in the application of recombinase-aided amplification combined with CRISPR-Cas technology in quick detection of pathogenic microbes.Front Bioeng Biotechnol. 2023 Aug 31;11:1215466. doi: 10.3389/fbioe.2023.1215466. eCollection 2023. Front Bioeng Biotechnol. 2023. PMID: 37720320 Free PMC article. Review.
-
RAP: A Novel Approach to the Rapid and Highly Sensitive Detection of Respiratory Viruses.Front Bioeng Biotechnol. 2021 Nov 5;9:766411. doi: 10.3389/fbioe.2021.766411. eCollection 2021. Front Bioeng Biotechnol. 2021. PMID: 34805120 Free PMC article. Review.
References
-
- Amalfitano N, Patel N, Haddi ML, Benabid H, Pazzola M, Vacca GM, et al Detailed mineral profile of milk, whey, and cheese from cows, buffaloes, goats, ewes, and dromedary camels, and efficiency of recovery of minerals in their cheese. J Dairy Sci. 2024;107(11):8887–907. doi: 10.3168/jds.2023-24624. - DOI - PubMed
-
- Ma XJ Isothermal amplification technology for diagnosis of COVID-19: current status and future prospects. Zoonoses. 2022;2(1):5. doi: 10.15212/zoonoses-2021-0022. - DOI
LinkOut - more resources
Full Text Sources
