A comprehensive multi-omics analysis uncovers the associations between gut microbiota and pancreatic cancer
- PMID: 40376462
- PMCID: PMC12078283
- DOI: 10.3389/fmicb.2025.1592549
A comprehensive multi-omics analysis uncovers the associations between gut microbiota and pancreatic cancer
Abstract
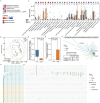
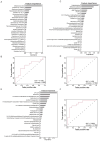
Pancreatic cancer is one of the most lethal malignant neoplasms. Pancreatic cancer is related to gut microbiota, but the associations between its treatment and microbial abundance as well as genetic variations remain unclear. In this study, we collected fecal samples from 58 pancreatic cancer patients including 43 pancreatic ductal adenocarcinoma (PDAC) and 15 non-PDAC, and 40 healthy controls, and shotgun metagenomic sequencing and untargeted metabolome analysis were conducted. PDAC patients were divided into five groups according to treatment and tumor location, including treatment-naive (UT), chemotherapy (CT), surgery combined with chemotherapy (SCT), Head, and body/tail (Tail) groups. Multivariate association analysis revealed that both CT and SCT were associated with increased abundance of Lactobacillus gasseri and Streptococcus equinus. The microbial single nucleotide polymorphisms (SNPs) densities of Streptococcus salivarius, Streptococcus vestibularis and Streptococcus thermophilus were positively associated with CT, while Lachnospiraceae bacterium 2_1_58FAA was positively associated with Head group. Compared with Tail group, the Head group showed positive associations with opportunistic pathogens, such as Escherichia coli, Shigella sonnei and Shigella flexneri. We assembled 424 medium-quality non-redundant metagenome-assembled genomes (nrMAGs) and 276 high-quality nrMAGs. In CT group, indole-3-acetic acid, capsaicin, sinigrin, chenodeoxycholic acid, and glycerol-3-phosphate were increased, and the accuracy of the model based on fecal metabolites reached 0.77 in distinguishing healthy controls and patients. This study identifies the associations between pancreatic cancer treatment and gut microbiota as well as its metabolites, reveals bacterial SNPs are related to tumor location, and extends our knowledge of gut microbiota and pancreatic cancer.
Keywords: gut microbiota; metabolome; metagenomic binning; pancreatic cancer; single nucleotide polymorphisms.
Copyright © 2025 Han, Cao, Tang and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Integrated analysis of microbiome and metabolome reveals signatures in PDAC tumorigenesis and prognosis.Microbiol Spectr. 2024 Nov 5;12(11):e0096224. doi: 10.1128/spectrum.00962-24. Epub 2024 Oct 10. Microbiol Spectr. 2024. PMID: 39387592 Free PMC article.
-
Metagenomic Study Reveals Phage-Bacterial Interactome Dynamics in Gut and Oral Microbiota in Pancreatic Diseases.Int J Mol Sci. 2024 Oct 12;25(20):10988. doi: 10.3390/ijms252010988. Int J Mol Sci. 2024. PMID: 39456772 Free PMC article.
-
Upregulation of Lactobacillus spp. in gut microbiota as a novel mechanism for environmental eustress-induced anti-pancreatic cancer effects.Gut Microbes. 2025 Dec;17(1):2470372. doi: 10.1080/19490976.2025.2470372. Epub 2025 Feb 23. Gut Microbes. 2025. PMID: 39988618 Free PMC article.
-
Metabolites derived from gut microbiota mitigate chemoresistance in pancreatic cancer.Expert Rev Gastroenterol Hepatol. 2024 Oct;18(10):597-604. doi: 10.1080/17474124.2024.2412045. Epub 2024 Oct 22. Expert Rev Gastroenterol Hepatol. 2024. PMID: 39439262 Review.
-
Gut microbiome changes associated with chronic pancreatitis and pancreatic cancer: a systematic review and meta-analysis.Int J Surg. 2024 Sep 1;110(9):5781-5794. doi: 10.1097/JS9.0000000000001724. Int J Surg. 2024. PMID: 38847785 Free PMC article.
References
-
- Altieri B., Grant W. B., Della Casa S., Orio F., Pontecorvi A., Colao A., et al. . (2017). Vitamin D and pancreas: the role of sunshine vitamin in the pathogenesis of diabetes mellitus and pancreatic cancer. Crit. Rev. Food Sci. Nutr. 57, 3472–3488. doi: 10.1080/10408398.2015.1136922, PMID: - DOI - PubMed
LinkOut - more resources
Full Text Sources

