Piperlongumine inhibits renal cell carcinoma progression by modulating the DDX11-miR-15b-3p-DLD axis
- PMID: 40376520
- PMCID: PMC12076233
- DOI: 10.21037/tau-2025-11
Piperlongumine inhibits renal cell carcinoma progression by modulating the DDX11-miR-15b-3p-DLD axis
Abstract
Background: Piperlongumine (PL) is a natural alkaloid obtained from the long pepper and can inhibit the progression of various tumors. However, its role in renal cell carcinoma (RCC) remains unclear. Thus, the purpose of this study was to determine whether PL can suppress RCC progression and to clarify the related mechanisms.
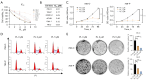
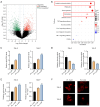
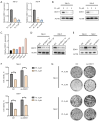
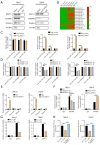
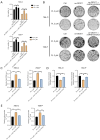
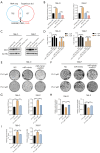
Methods: Cell Counting Kit-8 (CCK-8) and colony formation assays were applied to characterize the effect of PL in RCC cell proliferation; meanwhile, cellular reactive oxygen species (ROS), Fe2+ level, and mitochondrial membrane potential (MMP) assays were used to determine PL's role in mitochondrial homeostasis. Immunofluorescence experiments using MitoTracker red was applied to visualize the morphology of mitochondria. Western blotting and coimmunoprecipitation (Co-IP) and RNA immunoprecipitation (RNA-IP) assays were used to examine PL's regulation of DDX11 expression via miR-15b-3p. In addition, a xenograft mouse model was created to clarify the effect of DDX11 overexpression on RCC progression with or without PL treatment.
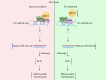
Results: We found that PL inhibited RCC cell proliferation in a dose-dependent manner by interfering with the mitochondrial homeostasis of RCC cells. In terms of mechanism, RNA sequencing showed that PL decreased the expression of DDX11, inhibited the maturation of miR-15b-3p, and further increased the level of dihydrolipoamide dehydrogenase (DLD) to disrupt the mitochondrial homeostasis of RCC cells. In addition, the vivo xenograft mouse model showed that DDX11 plays a stimulatory role in PL-induced RCC inhibition.
Conclusions: Our study demonstrated that DDX11 contributes to PL-induced RCC inhibition by modulating the miR-15b-3p-DLD axis.
Keywords: DDX11; Piperlongumine (PL); dihydrolipoamide dehydrogenase (DLD); miR-15b-3p; renal cell carcinoma (RCC).
Copyright © 2025 AME Publishing Company. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tau.amegroups.com/article/view/10.21037/tau-2025-11/coif). The authors have no conflicts of interest to declare.
Figures

Similar articles
-
Circ_0000274 contributes to renal cell carcinoma progression by regulating miR-338-3p/NUCB2 axis and JAK1/STAT3 pathway.Transpl Immunol. 2022 Oct;74:101626. doi: 10.1016/j.trim.2022.101626. Epub 2022 May 13. Transpl Immunol. 2022. PMID: 35569717
-
Long non-coding RNA DDX11-AS1 promotes esophageal carcinoma cell proliferation and migration through regulating the miR-514b-3p/RBX1 axis.Bioengineered. 2021 Dec;12(1):3772-3786. doi: 10.1080/21655979.2021.1940617. Bioengineered. 2021. PMID: 34281459 Free PMC article.
-
Silenced lncRNA DDX11-AS1 or up-regulated microRNA-34a-3p inhibits malignant phenotypes of hepatocellular carcinoma cells via suppression of TRAF5.Cancer Cell Int. 2021 Mar 22;21(1):179. doi: 10.1186/s12935-021-01847-6. Cancer Cell Int. 2021. PMID: 33752668 Free PMC article.
-
Piperlongumine and its analogs down-regulate expression of c-Met in renal cell carcinoma.Cancer Biol Ther. 2015;16(5):743-9. doi: 10.1080/15384047.2015.1026511. Cancer Biol Ther. 2015. PMID: 25801713 Free PMC article.
-
Piperlongumine inhibits head and neck squamous cell carcinoma proliferation by docking to Akt.Phytother Res. 2020 Dec;34(12):3345-3358. doi: 10.1002/ptr.6788. Epub 2020 Aug 15. Phytother Res. 2020. PMID: 32798277
References
LinkOut - more resources
Full Text Sources
Miscellaneous
