Development and validation of tryptophan metabolism-related risk model and molecular subtypes for predicting postoperative biochemical recurrence in prostate cancer
- PMID: 40376539
- PMCID: PMC12076229
- DOI: 10.21037/tau-2025-39
Development and validation of tryptophan metabolism-related risk model and molecular subtypes for predicting postoperative biochemical recurrence in prostate cancer
Abstract
Background: Biochemical recurrence (BCR) following radical prostatectomy (RP) remains a major challenge in prostate cancer (PCa) management. Tryptophan metabolism plays a pivotal role in tumor progression and immune modulation. This study aimed to develop and validate a tryptophan metabolism-related risk model and molecular subtypes to predict BCR in PCa patients after RP.
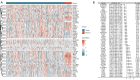
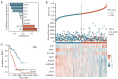
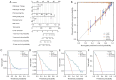
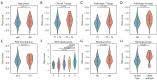
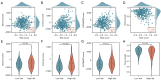
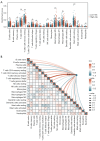
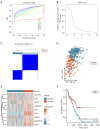
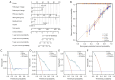
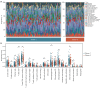
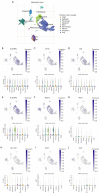
Methods: The Cancer Genome Atlas-Prostate Adenocarcinoma (TCGA-PRAD) dataset, including 421 PCa patients, was analyzed to identify key tryptophan metabolism-related genes (TMRGs) using differential expression, univariate Cox, and the least absolute shrinkage and selection operator (LASSO) regression analyses. The tryptophan metabolism-related risk model was constructed through multivariate Cox regression, and tryptophan metabolism-related molecular subtypes were established using consensus clustering. External validation was conducted using an independent dataset, while immunohistochemistry (IHC) and single-cell sequencing further confirmed TMRG expression patterns and their roles in the tumor microenvironment (TME).
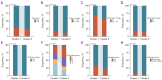
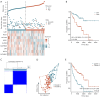
Results: The tryptophan metabolism-related risk model and molecular subtypes effectively stratified PCa patients into low- and high-risk groups or two molecular subtypes. High-risk PCa patients (n=211) and those in Cluster 1 (n=261) exhibited significantly poorer biochemical recurrence-free survival (BRFS) and distinct clinicopathological features, immune infiltration profiles, and TME characteristics. External validation confirmed the robustness of the tryptophan metabolism-related risk model and molecular subtypes. IHC and single-cell sequencing highlighted the expression patterns of TMRGs and their regulatory roles in the TME.
Conclusions: This study established and validated tryptophan metabolism-related risk scores and molecular subtypes as reliable predictors of BCR in PCa patients after RP. These findings provide a foundation for personalized follow-up and treatment strategies, contributing to improved clinical outcomes in PCa management.
Keywords: Prostate cancer (PCa); biochemical recurrence (BCR); molecular subtypes; risk model; tryptophan.
Copyright © 2025 AME Publishing Company. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tau.amegroups.com/article/view/10.21037/tau-2025-39/coif). The authors have no conflicts of interest to declare.
Figures


Similar articles
-
Identification of cuproptosis-related subtypes, construction of a prognosis model, and tumor microenvironment landscape in gastric cancer.Front Immunol. 2022 Nov 21;13:1056932. doi: 10.3389/fimmu.2022.1056932. eCollection 2022. Front Immunol. 2022. PMID: 36479114 Free PMC article.
-
A Novel Four Mitochondrial Respiration-Related Signature for Predicting Biochemical Recurrence of Prostate Cancer.J Clin Med. 2023 Jan 13;12(2):654. doi: 10.3390/jcm12020654. J Clin Med. 2023. PMID: 36675580 Free PMC article.
-
Identification and validation of a risk model and molecular subtypes based on tryptophan metabolism-related genes to predict the clinical prognosis and tumor immune microenvironment in lower-grade glioma.Front Cell Neurosci. 2023 Feb 28;17:1146686. doi: 10.3389/fncel.2023.1146686. eCollection 2023. Front Cell Neurosci. 2023. PMID: 36925967 Free PMC article.
-
Periprostatic fat magnetic resonance imaging based radiomics nomogram for predicting biochemical recurrence-free survival in patients with non-metastatic prostate cancer after radical prostatectomy.BMC Cancer. 2024 Nov 27;24(1):1459. doi: 10.1186/s12885-024-13207-4. BMC Cancer. 2024. PMID: 39604917 Free PMC article.
-
Development and validation of [18 F]-PSMA-1007 PET-based radiomics model to predict biochemical recurrence-free survival following radical prostatectomy.Eur J Nucl Med Mol Imaging. 2024 Jul;51(9):2806-2818. doi: 10.1007/s00259-024-06734-6. Epub 2024 May 1. Eur J Nucl Med Mol Imaging. 2024. PMID: 38691111
References
LinkOut - more resources
Full Text Sources
